Impaired degradation of PLCG1 by chaperone-mediated autophagy promotes cellular senescence and intervertebral disc degeneration
- PMID: 39212196
- PMCID: PMC11759519
- DOI: 10.1080/15548627.2024.2395797
Impaired degradation of PLCG1 by chaperone-mediated autophagy promotes cellular senescence and intervertebral disc degeneration
Abstract
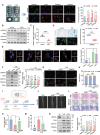
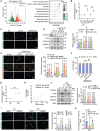
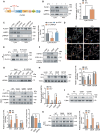
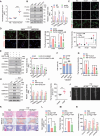
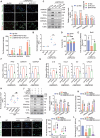
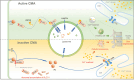
Defects in chaperone-mediated autophagy (CMA) are associated with cellular senescence, but the mechanism remains poorly understood. Here, we found that CMA inhibition induced cellular senescence in a calcium-dependent manner and identified its role in TNF-induced senescence of nucleus pulposus cells (NPC) and intervertebral disc degeneration. Based on structural and functional proteomic screens, PLCG1 (phospholipase C gamma 1) was predicted as a potential substrate for CMA deficiency to affect calcium homeostasis. We further confirmed that PLCG1 was a key mediator of CMA in the regulation of intracellular calcium flux. Aberrant accumulation of PLCG1 caused by CMA blockage resulted in calcium overload, thereby inducing NPC senescence. Immunoassays on human specimens showed that reduced LAMP2A, the rate-limiting protein of CMA, or increased PLCG1 was associated with disc senescence, and the TNF-induced disc degeneration in rats was inhibited by overexpression of Lamp2a or knockdown of Plcg1. Because CMA dysregulation, calcium overload, and cellular senescence are common features of disc degeneration and other age-related degenerative diseases, the discovery of actionable molecular targets that can link these perturbations may have therapeutic value.Abbreviation: ATRA: all-trans-retinoic acid; BrdU: bromodeoxyuridine; CDKN1A/p21: cyclin dependent kinase inhibitor 1A; CDKN2A/p16-INK4A: cyclin dependent kinase inhibitor 2A; CMA: chaperone-mediated autophagy; DHI: disc height index; ER: endoplasmic reticulum; IP: immunoprecipitation; IP3: inositol 1,4,5-trisphosphate; ITPR/IP3R: inositol 1,4,5-trisphosphate receptor; IVD: intervertebral disc; IVDD: intervertebral disc degeneration; KD: knockdown; KO: knockout; Leu: leupeptin; MRI: magnetic resonance imaging; MS: mass spectrometry; N/L: NH4Cl and leupeptin; NP: nucleus pulposus; NPC: nucleus pulposus cells; PI: protease inhibitors; PLC: phospholipase C; PLCG1: phospholipase C gamma 1; ROS: reactive oxygen species; RT-qPCR: real-time quantitative reverse transcription PCR; SA-GLB1/β-gal: senescence-associated galactosidase beta 1; SASP: senescence-associated secretory phenotype; STV: starvation; TMT: tandem mass tag; TNF: tumor necrosis factor; TP53: tumor protein p53; UPS: ubiquitin-proteasome system.
Keywords: Calcium overload; chaperone-mediated autophagy; intervertebral disc degeneration; nucleus pulposus; senescence.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
