RAC2 gain-of-function variants causing inborn error of immunity drive NLRP3 inflammasome activation
- PMID: 39212656
- PMCID: PMC11363864
- DOI: 10.1084/jem.20231562
RAC2 gain-of-function variants causing inborn error of immunity drive NLRP3 inflammasome activation
Abstract
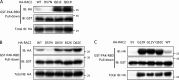
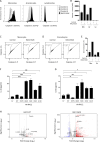
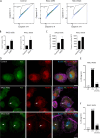
A growing number of patients presenting severe combined immunodeficiencies attributed to monoallelic RAC2 variants have been identified. The expression of the RHO GTPase RAC2 is restricted to the hematopoietic lineage. RAC2 variants have been described to cause immunodeficiencies associated with high frequency of infection, leukopenia, and autoinflammatory features. Here, we show that specific RAC2 activating mutations induce the NLRP3 inflammasome activation leading to the secretion of IL-1β and IL-18 from macrophages. This activation depends on the activation state of the RAC2 variant and is mediated by the downstream kinase PAK1. Inhibiting the RAC2-PAK1-NLRP3 inflammasome pathway might be considered as a potential treatment for these patients.
© 2024 Doye et al.
Conflict of interest statement
Disclosures: A. Doye, P. Chaintreuil, O. Visvikis, E. Verhoeyen, and L. Boyer reported a patent to EP23305994.8 pending and a patent to EP24306144.7 pending. S. Kracker reported a patent to EP23305994.8 pending. No other disclosures were reported.
Figures






References
-
- Accetta, D., Syverson G., Bonacci B., Reddy S., Bengtson C., Surfus J., Harbeck R., Huttenlocher A., Grossman W., Routes J., and Verbsky J.. 2011. Human phagocyte defect caused by a Rac2 mutation detected by means of neonatal screening for T-cell lymphopenia. J. Allergy Clin. Immunol. 127:535–538.e1: 2. 10.1016/j.jaci.2010.10.013 - DOI - PubMed
-
- Alkhairy, O.K., Rezaei N., Graham R.R., Abolhassani H., Borte S., Hultenby K., Wu C., Aghamohammadi A., Williams D.A., Behrens T.W., et al. . 2015. RAC2 loss-of-function mutation in 2 siblings with characteristics of common variable immunodeficiency. J. Allergy Clin. Immunol. 135:1380–1384.e1: 5. 10.1016/j.jaci.2014.10.039 - DOI - PMC - PubMed
-
- Ambruso, D.R., Knall C., Abell A.N., Panepinto J., Kurkchubasche A., Thurman G., Gonzalez-Aller C., Hiester A., deBoer M., Harbeck R.J., et al. . 2000. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc. Natl. Acad. Sci. USA. 97:4654–4659. 10.1073/pnas.080074897 - DOI - PMC - PubMed
-
- Batistic, L. and Boyer L.. 2024. RAC2 gain-of-function variants causing inborn error of immunity drive NLRP3 inflammasome activation. Dryad. 10.5061/dryad.p8cz8w9zx - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

