New insights into the biology of T-cell lymphomas
- PMID: 39213420
- PMCID: PMC11551850
- DOI: 10.1182/blood.2023021787
New insights into the biology of T-cell lymphomas
Abstract
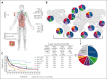
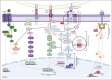
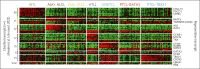
Peripheral T-cell lymphomas (PTCLs) encompass a heterogeneous group of postthymic T-cell lymphomas with >30 distinct subtypes associated with varied clinicopathological features. Unfortunately, the overall survival of the major PTCL subtypes is dismal and has not improved for decades; thus, there is an urgent unmet clinical need to improve diagnosis, therapies, and clinical outcomes. The diagnosis is often challenging, requiring a combinatorial evaluation of clinical, morphologic, and immunophenotypic features. PTCL pathobiology is difficult to investigate due to enormous intertumor and intratumor heterogeneity, limited tissue availability, and the paucity of authentic T-cell lymphoma cell lines or genetically faithful animal models. The application of transcriptomic profiling and genomic sequencing has markedly accelerated the discovery of new biomarkers, molecular signatures, and genetic lesions, and some of the discoveries have been included in the revised World Health Organization or International Consensus Classification. Genome-wide investigations have revealed the mutational landscape and transcriptomic profiles of PTCL entities, defined the cell of origin as a major determinant of T-cell lymphoma biology, and allowed for the refinement of biologically and clinically meaningful entities for precision therapy. In this review, we prioritize the discussion on common nodal PTCL subtypes together with 2 virus-associated T-cell and natural killer cell lymphomas. We succinctly review normal T-cell development, differentiation, and T-cell receptor signaling as they relate to PTCL pathogenesis and biology. This review will facilitate a better biological understanding of the different PTCL entities and their stratification for additional studies and target-directed clinical trials.
© 2024 American Society of Hematology. Published by Elsevier Inc. All rights are reserved, including those for text and data mining, AI training, and similar technologies.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



References
-
- Vose J, Armitage J, Weisenburger D, International T-Cell Lymphoma Project International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124–4130. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. WHO Classification of Tumors; 2017.
-
- Wang SS, Vose JM. In: T-Cell Lymphomas. Foss F, editor. Humana Press; 2013. Epidemiology and prognosis of T-cell lymphoma; pp. 25–39.
-
- Delabie J, Holte H, Vose JM, et al. Enteropathy-associated T-cell lymphoma: clinical and histological findings from the international peripheral T-cell lymphoma project. Blood. 2011;118(1):148–155. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

