Combination anti-PD-1 and anti-CTLA-4 therapy generates waves of clonal responses that include progenitor-exhausted CD8+ T cells
- PMID: 39214097
- PMCID: PMC11387127
- DOI: 10.1016/j.ccell.2024.08.007
Combination anti-PD-1 and anti-CTLA-4 therapy generates waves of clonal responses that include progenitor-exhausted CD8+ T cells
Abstract
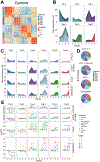
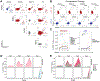
Combination checkpoint blockade with anti-PD-1 and anti-CTLA-4 antibodies has shown promising efficacy in melanoma. However, the underlying mechanism in humans remains unclear. Here, we perform paired single-cell RNA and T cell receptor (TCR) sequencing across time in 36 patients with stage IV melanoma treated with anti-PD-1, anti-CTLA-4, or combination therapy. We develop the algorithm Cyclone to track temporal clonal dynamics and underlying cell states. Checkpoint blockade induces waves of clonal T cell responses that peak at distinct time points. Combination therapy results in greater magnitude of clonal responses at 6 and 9 weeks compared to single-agent therapies, including melanoma-specific CD8+ T cells and exhausted CD8+ T cell (TEX) clones. Focused analyses of TEX identify that anti-CTLA-4 induces robust expansion and proliferation of progenitor TEX, which synergizes with anti-PD-1 to reinvigorate TEX during combination therapy. These next generation immune profiling approaches can guide the selection of drugs, schedule, and dosing for novel combination strategies.
Keywords: CTLA-4 blockade; Checkpoint blockade; PD-1 blockade; T cell exhaustion; clonotypic analysis; combination checkpoint blockade; immunotherapy; melanoma; progenitor exhausted CD8(+) T cells; single-cell sequencing.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.C.H. performed consulting work for Immunai and received research funding from Bristol Myers Squibb and Merck. R.S.H. has performed consulting work for Bristol Myers Squibb (exclusive of the current work). T.C.M. received honorarium for Scientific Advisory Board participation from: BMS, GigaGen, Merck, Pliant, Pfizer. G.C.K. is on the Merck Advisory Board. J.W. consulted for and have received less than $10,000 per annum from Merck, Genentech, AstraZeneca, GSK, Novartis, Nektar, Celldex, Incyte, Biond, Moderna, ImCheck, Sellas, Evaxion, Pfizer, Regeneron, and EMD Serono and received $10–$25,000 from BMS for membership on advisory boards. J.W. also holds equity in Biond, Evaxion, OncoC4, and Instil Bio, and on scientific advisory boards for CytomX, Incyte, ImCheck, Biond, Sellas, Instil Bio, OncoC4, and NexImmune and remunerated between $10,000–$50,000. In addition, J.W. is named on a patent filed by Moffitt Cancer Center on an ipilimumab biomarker and on TIL preparation and also on a PD-1 patent filed by Biodesix; J.W. receives less than $6000 in royalties. D.B., E.K., and C.S. were employed by Immunai when engaged in this project. S.G. and D.T. are employees of BMS. C.A. is a consultant for Biotherapy Partners.
Figures



References
-
- Wei SC, Duffy CR, and Allison JP (2018). Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov 8, 1069–1086. 10.1158/2159-8290.CD-18-0367. - DOI - PubMed
-
- Brunner MC, Chambers CA, Chan FK, Hanke J, Winoto A, and Allison JP (1999). CTLA-4-Mediated inhibition of early events of T cell proliferation. J Immunol 162, 5813–5820. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

