Sde proteins coordinate ubiquitin utilization and phosphoribosylation to establish and maintain the Legionella replication vacuole
- PMID: 39214970
- PMCID: PMC11364549
- DOI: 10.1038/s41467-024-51272-2
Sde proteins coordinate ubiquitin utilization and phosphoribosylation to establish and maintain the Legionella replication vacuole
Abstract
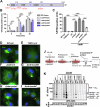
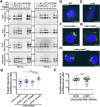
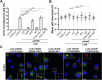
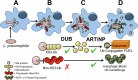
The Legionella pneumophila Sde family of translocated proteins promotes host tubular endoplasmic reticulum (ER) rearrangements that are tightly linked to phosphoribosyl-ubiquitin (pR-Ub) modification of Reticulon 4 (Rtn4). Sde proteins have two additional activities of unclear relevance to the infection process: K63 linkage-specific deubiquitination and phosphoribosyl modification of polyubiquitin (pR-Ub). We show here that the deubiquitination activity (DUB) stimulates ER rearrangements while pR-Ub protects the replication vacuole from cytosolic surveillance by autophagy. Loss of DUB activity is tightly linked to lowered pR-Ub modification of Rtn4, consistent with the DUB activity fueling the production of pR-Ub-Rtn4. In parallel, phosphoribosyl modification of polyUb, in a region of the protein known as the isoleucine patch, prevents binding by the autophagy adapter p62. An inability of Sde mutants to modify polyUb results in immediate p62 association, a critical precursor to autophagic attack. The ability of Sde WT to block p62 association decays quickly after bacterial infection, as predicted by the presence of previously characterized L. pneumophila effectors that inactivate Sde and remove polyUb. In sum, these results show that the accessory Sde activities act to stimulate ER rearrangements and protect from host innate immune sensing in a temporal fashion.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Sde Proteins Coordinate Ubiquitin Utilization and Phosphoribosylation to Promote Establishment and Maintenance of the Legionella Replication Vacuole.Res Sq [Preprint]. 2023 Sep 18:rs.3.rs-3269310. doi: 10.21203/rs.3.rs-3269310/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Aug 30;15(1):7479. doi: 10.1038/s41467-024-51272-2. PMID: 37790456 Free PMC article. Updated. Preprint.
-
Sde Proteins Coordinate Ubiquitin Utilization and Phosphoribosylation to Establish and Maintain the Legionella Replication Vacuole.bioRxiv [Preprint]. 2024 Jul 16:2023.09.07.553534. doi: 10.1101/2023.09.07.553534. bioRxiv. 2024. Update in: Nat Commun. 2024 Aug 30;15(1):7479. doi: 10.1038/s41467-024-51272-2. PMID: 38645023 Free PMC article. Updated. Preprint.
References
-
- Safdar, N., Crnich, C. J. & Maki, D. G. The pathogenesis of ventilator-associated pneumonia: its relevance to developing effective strategies for prevention. Respir. Care50, 725–739 (2005). - PubMed
-
- Horwitz, M. A. & Silverstein, S. C. Interaction of the Legionnaires’ disease bacterium (Legionella pneumophila) with human phagocytes. I. L. pneumophila resists killing by polymorphonuclear leukocytes, antibody, and complement. J. Exp. Med. 153, 386–397 (1981). 10.1084/jem.153.2.386 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- T32 GM007310/GM/NIGMS NIH HHS/United States
- R01AI113211/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01AI146245/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI113211/AI/NIAID NIH HHS/United States
- R01 GM132422/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

