Mapping the immunopeptidome of seven SARS-CoV-2 antigens across common HLA haplotypes
- PMID: 39214998
- PMCID: PMC11364864
- DOI: 10.1038/s41467-024-51959-6
Mapping the immunopeptidome of seven SARS-CoV-2 antigens across common HLA haplotypes
Abstract
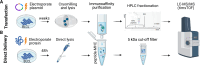
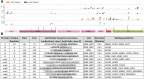
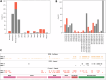
Most COVID-19 vaccines elicit immunity against the SARS-CoV-2 Spike protein. However, Spike protein mutations in emerging strains and immune evasion by the SARS-CoV-2 virus demonstrates the need to develop more broadly targeting vaccines. To facilitate this, we use mass spectrometry to identify immunopeptides derived from seven relatively conserved structural and non-structural SARS-CoV-2 proteins (N, E, Nsp1/4/5/8/9). We use two different B-lymphoblastoid cell lines to map Human Leukocyte Antigen (HLA) class I and class II immunopeptidomes covering some of the prevalent HLA types across the global human population. We employ DNA plasmid transfection and direct antigen delivery approaches to sample different antigens and find 248 unique HLA class I and HLA class II bound peptides with 71 derived from N, 12 from E, 28 from Nsp1, 19 from Nsp4, 73 from Nsp8 and 45 peptides derived from Nsp9. Over half of the viral peptides are unpublished. T cell reactivity tested against 56 of the detected peptides shows CD8+ and CD4+ T cell responses against several peptides from the N, E, and Nsp9 proteins. Results from this study will aid the development of next-generation COVID vaccines targeting epitopes from across a number of SARS-CoV-2 proteins.
© 2024. The Author(s).
Conflict of interest statement
NT, EJL, GP, MSK, JVK, and ABS are employed by Evaxion Biotech A/S which holds IP for identifying neoepitopes and personalized immunotherapy. AWP is on the scientific advisory board of Evaxion Biotech A/S. AWP is on the advisory board of Bioinformatics Solutions (Canada), and Grey Wolf Therapeutics (UK) and is a co-founder of Resseptor Therapeutics (Australia). The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- 2016596/Department of Health | National Health and Medical Research Council (NHMRC)
- 1143366/Department of Health | National Health and Medical Research Council (NHMRC)
- 1173871/Department of Health | National Health and Medical Research Council (NHMRC)
- 1194036/Department of Health | National Health and Medical Research Council (NHMRC)
- 2026357/Department of Health | National Health and Medical Research Council (NHMRC)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

