RAS-ON inhibition overcomes clinical resistance to KRAS G12C-OFF covalent blockade
- PMID: 39215000
- PMCID: PMC11364849
- DOI: 10.1038/s41467-024-51828-2
RAS-ON inhibition overcomes clinical resistance to KRAS G12C-OFF covalent blockade
Abstract
Selective KRASG12C inhibitors have been developed to covalently lock the oncogene in the inactive GDP-bound state. Two of these molecules, sotorasib and adagrasib, are approved for the treatment of adult patients with KRASG12C-mutated previously treated advanced non-small cell lung cancer. Drug treatment imposes selective pressures leading to the outgrowth of drug-resistant variants. Mass sequencing from patients' biopsies identified a number of acquired KRAS mutations -both in cis and in trans- in resistant tumors. We demonstrate here that disease progression in vivo can also occur due to adaptive mechanisms and increased KRAS-GTP loading. Using the preclinical tool tri-complex KRASG12C-selective covalent inhibitor, RMC-4998 (also known as RM-029), that targets the active GTP-bound (ON) state of the oncogene, we provide a proof-of-concept that the clinical stage KRASG12C(ON) inhibitor RMC-6291 alone or in combination with KRASG12C(OFF) drugs can be an alternative potential therapeutic strategy to circumvent resistance due to increased KRAS-GTP loading.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: D.S. received research fees from Aelin Therapeutics. C.A. received research fees from Revolution Medicines, Aelin Therapeutics, Verastem, Roche, and Boehringer Ingelheim. A.I. received research support from AstraZeneca, Bayer, BMS, Merck, MSD, Pharmamar, and Roche and participated as a consultant in advisory boards for Bayer, Blueprint, Daiichi Sankyo, Epizyme, Fstar Therapeutics, Roche and Springworks. S.C. participated as a consultant on advisory boards for Roche, BMS, and AstraZeneca. B.R. received advisory board/consulting and honoraria fees from AstraZeneca, Regeneron, Amgen, Bayer, SITC, and Targeted Oncology. M.M.A. received advisory board/consulting and research fees from Merck, Pfizer, Bristol Myers Squibb, Foundation Medicine, Novartis, Gritstone Bio, Mirati Therapeutics, EMD Serono, AstraZeneca, Instil Bio, Regeneron, Janssen, Affini-T Therapeutics, Genentech/Roche, Lilly, Amgen. E.N. received research support from Roche, Pfizer, Bristol Myers Squibb and Merck Serono and participated as consultant in advisory boards for Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo, Janssen, Lilly, Merck Serono, Merck Sharpe & Dohme, Pfizer, Roche, Sanofi and Takeda. E.N. participated as an investigator of clinical trials from Amgen and Revolution Medicines related to the current publication. Au.V. and A.V. are founders of the spin-off Xenopat S.L. The remaining authors declare no competing interests.
Figures


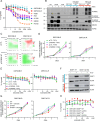
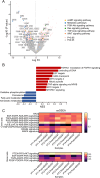

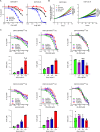


References
-
- Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer20, 555–572 (2020). - PubMed
-
- Canon, J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature575, 217–223 (2019). - PubMed
-
- Riely, G. J. et al. 99O_PR KRYSTAL-1: activity and preliminary pharmacodynamic (PD) analysis of adagrasib (MRTX849) in patients (Pts) with advanced non–small cell lung cancer (NSCLC) harboring KRASG12C mutation. J. Thorac. Oncol. 16, S751–S752 (2021).
MeSH terms
Substances
Grants and funding
- CDA-2019/Giovanni Armenise-Harvard Foundation
- 101001288/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- IG 2021 - ID. 25737/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- PI21/00789, PI21/00789, FIS PI22-00548/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- PI19/01320/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- 2021SGR00184/Generalitat de Catalunya (Government of Catalonia)
- MFAG 2017 ID. 20566, BRIDGE 2023 - ID. 28739/Fondazione Italiana per la Ricerca sul Cancro (Italian Foundation for Cancer Research)
- INCA_13911/Institut National Du Cancer (French National Cancer Institute)
- EPAEC222641CICS, PRYGN222960SANT/Fundación Científica Asociación Española Contra el Cáncer (Scientific Foundation, Spanish Association Against Cancer)

