Direct inhibition of tumor hypoxia response with synthetic transcriptional repressors
- PMID: 39215099
- PMCID: PMC12206482
- DOI: 10.1038/s41589-024-01716-z
Direct inhibition of tumor hypoxia response with synthetic transcriptional repressors
Abstract
Many oncogenic transcription factors (TFs) are considered to be undruggable because of their reliance on large protein-protein and protein-DNA interfaces. TFs such as hypoxia-inducible factors (HIFs) and X-box-binding protein 1 (XBP1) are induced by hypoxia and other stressors in solid tumors and bind to unfolded protein response element (UPRE) and hypoxia-induced response element (HRE) motifs to control oncogenic gene programs. Here, we report a strategy to create synthetic transcriptional repressors (STRs) that mimic the basic leucine zipper domain of XBP1 and recognize UPRE and HRE motifs. A lead molecule, STR22, binds UPRE and HRE DNA sequences with high fidelity and competes with both TFs in cells. Under hypoxia, STR22 globally suppresses HIF1α binding to HRE-containing promoters and enhancers, inhibits hypoxia-induced gene expression and blocks protumorigenic phenotypes in triple-negative breast cancer (TNBC) cells. In vivo, intratumoral and systemic STR22 treatment inhibited hypoxia-dependent gene expression, primary tumor growth and metastasis of TNBC tumors. These data validate a novel strategy to target the tumor hypoxia response through coordinated inhibition of TF-DNA binding.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: Z.Q., L.C.N., C.S.S., S.A.O., M.R.R. and R.E.M. are listed as inventors on patent (PCT/US22/80387) applications related to this study. S.A.O. is a cofounder, equity holder and consultant for OptiKIRA, LLC. R.E.M. is a cofounder and consultant for Anastasis Biotec, Ltd. and cofounder, director and consultant for ReAx Biotechnologies, Inc. All other authors declare no competing interests.
Figures



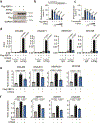


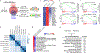
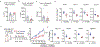





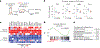
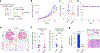
References
-
- Gatenby RA & Gillies RJ Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 (2004). - PubMed
-
- Hanahan D. & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
-
- Kim JW, Gao P. & Dang CV Effects of hypoxia on tumor metabolism. Cancer Metastasis Rev. 26, 291–298 (2007). - PubMed
-
- Liao D. & Johnson RS Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev. 26, 281–290 (2007). - PubMed
MeSH terms
Substances
Grants and funding
- R01 EY027810/EY/NEI NIH HHS/United States
- R01 CA219815/CA/NCI NIH HHS/United States
- RSG-17-150-01-CDD/American Cancer Society (American Cancer Society, Inc.)
- R01 GM145852/GM/NIGMS NIH HHS/United States
- R01 CA292876/CA/NCI NIH HHS/United States
- DP2GM128199; R01GM145852; F32GM148062/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 CA266643/CA/NCI NIH HHS/United States
- W81XWH2110120/U.S. Department of Defense (United States Department of Defense)
- P30 CA014599/CA/NCI NIH HHS/United States
- F32 GM148062/GM/NIGMS NIH HHS/United States
- R01EY027810; R01CA219815; R01CA266643;/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- DP2 GM128199/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

