µMap proximity labeling in living cells reveals stress granule disassembly mechanisms
- PMID: 39215100
- PMCID: PMC11868469
- DOI: 10.1038/s41589-024-01721-2
µMap proximity labeling in living cells reveals stress granule disassembly mechanisms
Abstract
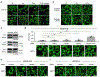
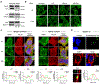
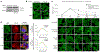
Phase-separated condensates are membrane-less intracellular structures comprising dynamic protein interactions that organize essential biological processes. Understanding the composition and dynamics of these organelles advances our knowledge of cellular behaviors and disease pathologies related to granule dysregulation. In this study, we apply microenvironment mapping with a HaloTag-based platform (HaloMap) to characterize intracellular stress granule dynamics in living cells. After validating the robustness and sensitivity of this approach, we then profile the stress granule proteome throughout the formation and disassembly and under pharmacological perturbation. These experiments reveal several ubiquitin-related modulators, including the HECT (homologous to E6AP C terminus) E3 ligases ITCH and NEDD4L, as well as the ubiquitin receptor toll-interacting protein TOLLIP, as key mediators of granule disassembly. In addition, we identify an autophagy-related pathway that promotes granule clearance. Collectively, this work establishes a general photoproximity labeling approach for unraveling intracellular protein interactomes and uncovers previously unexplored regulatory mechanisms of stress granule dynamics.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: D.W.C.M. declares an ownership interest in the company Dexterity Pharma, which has commercialized materials used in this work. D.W.C.M., C.P., S.D.K. and S.W.H. are co-inventors on a patent application (PCT/US23/79482) filed by Princeton University based on the HaloMap technology covered in this article.
Figures






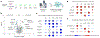


References
MeSH terms
Substances
Grants and funding
- 1F32GM142206-01/U.S. Department of Health & Human Services | NIH | Office of Extramural Research, National Institutes of Health (OER)
- F32 GM142206/GM/NIGMS NIH HHS/United States
- R35GM134897/U.S. Department of Health & Human Services | NIH | Office of Extramural Research, National Institutes of Health (OER)
- K99 GM154140/GM/NIGMS NIH HHS/United States
- R35 GM134897/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

