Distinct biological signature and modifiable risk factors underlie the comorbidity between major depressive disorder and cardiovascular disease
- PMID: 39215135
- PMCID: PMC11182748
- DOI: 10.1038/s44161-024-00488-y
Distinct biological signature and modifiable risk factors underlie the comorbidity between major depressive disorder and cardiovascular disease
Abstract
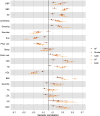
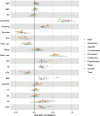
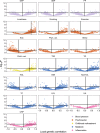
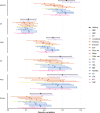
Major depressive disorder (MDD) and cardiovascular disease (CVD) are often comorbid, resulting in excess morbidity and mortality. Here we show that CVDs share most of their genetic risk factors with MDD. Multivariate genome-wide association analysis of shared genetic liability between MDD and atherosclerotic CVD revealed seven loci and distinct patterns of tissue and brain cell-type enrichments, suggesting the involvement of the thalamus. Part of the genetic overlap was explained by shared inflammatory, metabolic and psychosocial or lifestyle risk factors. Our data indicated causal effects of genetic liability to MDD on CVD risk, but not from most CVDs to MDD, and showed that the causal effects were partly explained by metabolic and psychosocial or lifestyle factors. The distinct signature of MDD-atherosclerotic CVD comorbidity suggests an immunometabolic subtype of MDD that is more strongly associated with CVD than overall MDD. In summary, we identified biological mechanisms underlying MDD-CVD comorbidity and modifiable risk factors for prevention of CVD in individuals with MDD.
© 2024. The Author(s).
Conflict of interest statement
C.M.L. sits on the SAB of Myriad Neuroscience and has received speaker/consultancy fees from SYNLAB and UCB. A.D.B. has received speaker’s honoraria from Lundbeck. O.A.A. is a consultant to Cortechs.ai and Precision Health, and has received speaker’s honoraria from Lundbeck, Janssen, Otsuka and Sunovion. P.F.S. has received consulting fees from and is a shareholder of Neumora Therapeutics. The other authors declare no competing interests.
Figures


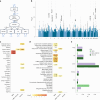
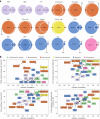


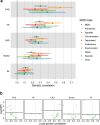

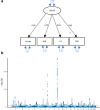

Update of
-
Distinct genomic signatures and modifiable risk factors underly the comorbidity between major depressive disorder and cardiovascular disease.medRxiv [Preprint]. 2024 Jan 29:2023.09.01.23294931. doi: 10.1101/2023.09.01.23294931. medRxiv. 2024. Update in: Nat Cardiovasc Res. 2024 Jun;3(6):754-769. doi: 10.1038/s44161-024-00488-y. PMID: 37693619 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
- R01 MH124851/MH/NIMH NIH HHS/United States
- MH123724/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- 220857/Z/20/Z/Wellcome Trust (Wellcome)
- 847776/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- R01 MH123724/MH/NIMH NIH HHS/United States
- MH121545/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- MH124871/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R155-2014-1724/Lundbeckfonden (Lundbeck Foundation)
- R01 MH124871/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 801133/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Sklodowska-Curie Actions (H2020 Excellent Science - Marie Sklodowska-Curie Actions)
- 964874/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- R01 MH121545/MH/NIMH NIH HHS/United States
- R01MH123724/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- D0886501/Vetenskapsrådet (Swedish Research Council)
LinkOut - more resources
Full Text Sources
