Writers, readers, and erasers RNA modifications and drug resistance in cancer
- PMID: 39215288
- PMCID: PMC11363509
- DOI: 10.1186/s12943-024-02089-6
Writers, readers, and erasers RNA modifications and drug resistance in cancer
Abstract
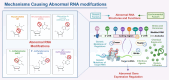
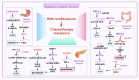
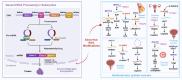
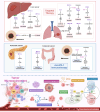
Drug resistance in cancer cells significantly diminishes treatment efficacy, leading to recurrence and metastasis. A critical factor contributing to this resistance is the epigenetic alteration of gene expression via RNA modifications, such as N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), 7-methylguanosine (m7G), pseudouridine (Ψ), and adenosine-to-inosine (A-to-I) editing. These modifications are pivotal in regulating RNA splicing, translation, transport, degradation, and stability. Governed by "writers," "readers," and "erasers," RNA modifications impact numerous biological processes and cancer progression, including cell proliferation, stemness, autophagy, invasion, and apoptosis. Aberrant RNA modifications can lead to drug resistance and adverse outcomes in various cancers. Thus, targeting RNA modification regulators offers a promising strategy for overcoming drug resistance and enhancing treatment efficacy. This review consolidates recent research on the role of prevalent RNA modifications in cancer drug resistance, with a focus on m6A, m1A, m5C, m7G, Ψ, and A-to-I editing. Additionally, it examines the regulatory mechanisms of RNA modifications linked to drug resistance in cancer and underscores the existing limitations in this field.
Keywords: Abnormal expression; Combination therapy; Drug resistance; Epigenetic alterations; RNA modification regulators.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

