Synthetic lethality of combined ULK1 defection and p53 restoration induce pyroptosis by directly upregulating GSDME transcription and cleavage activation through ROS/NLRP3 signaling
- PMID: 39215364
- PMCID: PMC11363528
- DOI: 10.1186/s13046-024-03168-8
Synthetic lethality of combined ULK1 defection and p53 restoration induce pyroptosis by directly upregulating GSDME transcription and cleavage activation through ROS/NLRP3 signaling
Abstract
Background: High expression of ubiquitin ligase MDM2 is a primary cause of p53 inactivation in many tumors, making it a promising therapeutic target. However, MDM2 inhibitors have failed in clinical trials due to p53-induced feedback that enhances MDM2 expression. This underscores the urgent need to find an effective adaptive genotype or combination of targets.
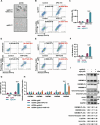
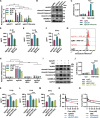
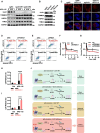
Methods: Kinome-wide CRISPR/Cas9 knockout screen was performed to identify genes that modulate the response to MDM2 inhibitor using TP53 wild type cancer cells and found ULK1 as a candidate. The MTT cell viability assay, flow cytometry and LDH assay were conducted to evaluate the activation of pyroptosis and the synthetic lethality effects of combining ULK1 depletion with p53 activation. Dual-luciferase reporter assay and ChIP-qPCR were performed to confirm that p53 directly mediates the transcription of GSDME and to identify the binding region of p53 in the promoter of GSDME. ULK1 knockout / overexpression cells were constructed to investigate the functional role of ULK1 both in vitro and in vivo. The mechanism of ULK1 depletion to activate GSMDE was mainly investigated by qPCR, western blot and ELISA.
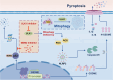
Results: By using high-throughput screening, we identified ULK1 as a synthetic lethal gene for the MDM2 inhibitor APG115. It was determined that deletion of ULK1 significantly increased the sensitivity, with cells undergoing typical pyroptosis. Mechanistically, p53 promote pyroptosis initiation by directly mediating GSDME transcription that induce basal-level pyroptosis. Moreover, ULK1 depletion reduces mitophagy, resulting in the accumulation of damaged mitochondria and subsequent increasing of reactive oxygen species (ROS). This in turn cleaves and activates GSDME via the NLRP3-Caspase inflammatory signaling axis. The molecular cascade makes ULK1 act as a crucial regulator of pyroptosis initiation mediated by p53 activation cells. Besides, mitophagy is enhanced in platinum-resistant tumors, and ULK1 depletion/p53 activation has a synergistic lethal effect on these tumors, inducing pyroptosis through GSDME directly.
Conclusion: Our research demonstrates that ULK1 deficiency can synergize with MDM2 inhibitors to induce pyroptosis. p53 plays a direct role in activating GSDME transcription, while ULK1 deficiency triggers upregulation of the ROS-NLRP3 signaling pathway, leading to GSDME cleavage and activation. These findings underscore the pivotal role of p53 in determining pyroptosis and provide new avenues for the clinical application of p53 restoration therapies, as well as suggesting potential combination strategies.
Keywords: MDM2 inhibitor; Mitophagy; Pyroptosis; Reactive oxygen species; TP53; ULK1.
© 2024. The Author(s).
Conflict of interest statement
All authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

