Bridging the gap: Multi-omics profiling of brain tissue in Alzheimer's disease and older controls in multi-ethnic populations
- PMID: 39215503
- PMCID: PMC11485084
- DOI: 10.1002/alz.14208
Bridging the gap: Multi-omics profiling of brain tissue in Alzheimer's disease and older controls in multi-ethnic populations
Abstract
Introduction: Multi-omics studies in Alzheimer's disease (AD) revealed many potential disease pathways and therapeutic targets. Despite their promise of precision medicine, these studies lacked Black Americans (BA) and Latin Americans (LA), who are disproportionately affected by AD.
Methods: To bridge this gap, Accelerating Medicines Partnership in Alzheimer's Disease (AMP-AD) expanded brain multi-omics profiling to multi-ethnic donors.
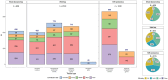
Results: We generated multi-omics data and curated and harmonized phenotypic data from BA (n = 306), LA (n = 326), or BA and LA (n = 4) brain donors plus non-Hispanic White (n = 252) and other (n = 20) ethnic groups, to establish a foundational dataset enriched for BA and LA participants. This study describes the data available to the research community, including transcriptome from three brain regions, whole genome sequence, and proteome measures.
Discussion: The inclusion of traditionally underrepresented groups in multi-omics studies is essential to discovering the full spectrum of precision medicine targets that will be pertinent to all populations affected with AD.
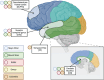
Highlights: Accelerating Medicines Partnership in Alzheimer's Disease Diversity Initiative led brain tissue profiling in multi-ethnic populations. Brain multi-omics data is generated from Black American, Latin American, and non-Hispanic White donors. RNA, whole genome sequencing and tandem mass tag proteomicsis completed and shared. Multiple brain regions including caudate, temporal and dorsolateral prefrontal cortex were profiled.
Keywords: Alzheimer's disease; data descriptor; multi‐omics; precision medicine; proteome; transcriptome; whole genome sequencing.
© 2024 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
The authors declare no conflicts of interest. Author disclosures are available in the supporting information.
Figures
Update of
-
Bridging the Gap: Multi-Omics Profiling of Brain Tissue in Alzheimer's Disease and Older Controls in Multi-Ethnic Populations.bioRxiv [Preprint]. 2024 Apr 20:2024.04.16.589592. doi: 10.1101/2024.04.16.589592. bioRxiv. 2024. Update in: Alzheimers Dement. 2024 Oct;20(10):7174-7192. doi: 10.1002/alz.14208. PMID: 38659743 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- R01 AG025711/AG/NIA NIH HHS/United States
- Michael J. Fox Foundation for Parkinson's Research
- RF1 AG062181/AG/NIA NIH HHS/United States
- R01 AG072120/AG/NIA NIH HHS/United States
- Mayo Foundation
- P30 AG072980/AG/NIA NIH HHS/United States
- R01-NS080820/NS/NINDS NIH HHS/United States
- P30AG066511/AG/NIA NIH HHS/United States
- U01 AG061357/AG/NIA NIH HHS/United States
- R01 AG003949/AG/NIA NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- P30AG072979/AG/NIA NIH HHS/United States
- R01 AG017216/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- CurePSP Foundation
- P50 AG016574/AG/NIA NIH HHS/United States
- P30 AG066511/AG/NIA NIH HHS/United States
- U19 AG074879/AG/NIA NIH HHS/United States
- P30AG072980/National Brain and Tissue Resource for Parkinson's Disease and Related Disorders
- U24 AG061340/AG/NIA NIH HHS/United States
- P01AG066597/AG/NIA NIH HHS/United States
- U19AG062418/AG/NIA NIH HHS/United States
- Alzheimer's Association Zenith Fellows Award
- Arizona Department of Health Services
- R01 AG061796/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- P01 AG066597/AG/NIA NIH HHS/United States
- U01 AG006576/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- R01 AG022018/AG/NIA NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- U19 AG062418/AG/NIA NIH HHS/United States
- U01AG061357/AG/NIA NIH HHS/United States
- Arizona Biomedical Research Commission
- U01 AG088425/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
- R01 AG075827/AG/NIA NIH HHS/United States
- RF1AG062181/AG/NIA NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- R01 AG072474/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG079170/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- P30 AG072979/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

