A new 1,2,3-triazole-indirubin hybrid suppresses tumor growth and pulmonary metastasis by mitigating the HGF/c-MET axis in hepatocellular carcinoma
- PMID: 39216686
- PMCID: PMC12225912
- DOI: 10.1016/j.jare.2024.08.033
A new 1,2,3-triazole-indirubin hybrid suppresses tumor growth and pulmonary metastasis by mitigating the HGF/c-MET axis in hepatocellular carcinoma
Abstract
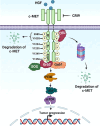
Introduction: Hepatocellular carcinoma (HCC) is a fatal cancer that is often diagnosed at the advanced stages which limits the available therapeutic options. The interaction of HGF with c-MET (a receptor tyrosine kinase) results in the activation of c-MET which subsequently triggers the PI3K/Akt/mTOR axis. Overexpression of c-MET in HCC tissues has been demonstrated to contribute to tumor progression and metastasis.
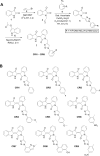
Objectives: We aimed to synthesize triazole-indirubin conjugates, examine their growth suppressor efficacy in cell-based assays, and investigate the antitumor as well as antimetastatic activity of lead cytotoxic agent in the orthotopic mice model.
Methods: A series of triazole-indirubin hybrids were synthesized and cytotoxicity, apoptogenic, and antimigratory effect of the lead compound (CRI9) was evaluated using MTT assay, cell cycle analysis, annexin-V/PI assay, TUNEL assay, and wound healing assay. The effect of CRI9 on the operation of the HGF/c-MET/PI3K/Akt/mTOR axis was examined using western blotting and transfection experiments. Acute toxicity, antitumor, and antimetastatic activity of CRI9 were examined in NCr nude mice. The expression of c-MET/PI3K/Akt/mTOR, CD31, and Ki-67 was examined using immunohistochemistry and western blotting.
Results: Among the new compounds, CRI9 consistently displayed potent cytotoxicity against HGF-induced HCC cells. CRI9 induced apoptosis as evidenced by increased sub G1 cells, annexin-V+/PI+ cells, TUNEL+ cells, and cleavage of procaspase-3 and PARP. CRI9 inhibited HGF-induced phosphorylation of c-METY1234/1235 and subsequently suppressed the PI3K/Akt/mTOR axis. Also, depletion of c-MET or inhibition of c-MET by CRI9 resulted in suppression of the PI3K/Akt/mTOR axis. CRI9 showed no toxic effects in NCr nude mice and displayed a potent antitumor and antimetastatic effect in the orthotopic HCC mice model. CRI9 also reduced the levels of phospho-c-MET, CD31, and Ki-67 and suppressed the activation of the PI3K/Akt/mTOR axis in tumor tissues.
Conclusion: CRI9 has been identified as a new inhibitor of the c-MET/PI3K/Akt/mTOR axis in HCC preclinical models.
Keywords: Hepatocellular carcinoma; Indirubin; Orthotopic mice model; Triazole; c-MET.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Mohan C.D., Bharathkumar H., Bulusu K.C., Pandey V., Rangappa S., Fuchs J.E., et al. Development of a novel azaspirane that targets the Janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo. J Biol Chem. 2014;289(49):34296–34307. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

