PARP1-TRIM44-MRN loop dictates the response to PARP inhibitors
- PMID: 39217466
- PMCID: PMC11514498
- DOI: 10.1093/nar/gkae756
PARP1-TRIM44-MRN loop dictates the response to PARP inhibitors
Abstract
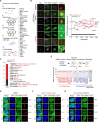
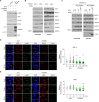
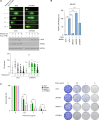
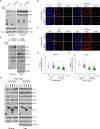
PARP inhibitors (PARPi) show selective efficacy in tumors with homologous recombination repair (HRR)-defects but the activation mechanism of HRR pathway in PARPi-treated cells remains enigmatic. To unveil it, we searched for the mediator bridging PARP1 to ATM pathways by screening 211 human ubiquitin-related proteins. We discovered TRIM44 as a crucial mediator that recruits the MRN complex to damaged chromatin, independent of PARP1 activity. TRIM44 binds PARP1 and regulates the ubiquitination-PARylation balance of PARP1, which facilitates timely recruitment of the MRN complex for DSB repair. Upon exposure to PARPi, TRIM44 shifts its binding from PARP1 to the MRN complex via its ZnF UBP domain. Knockdown of TRIM44 in cells significantly enhances the sensitivity to olaparib and overcomes the resistance to olaparib induced by 53BP1 deficiency. These observations emphasize the central role of TRIM44 in tethering PARP1 to the ATM-mediated repair pathway. Suppression of TRIM44 may enhance PARPi effectiveness and broaden their use even to HR-proficient tumors.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

