Evaluation of the effect of RNA secondary structure on Cas13d-mediated target RNA cleavage
- PMID: 39220269
- PMCID: PMC11364014
- DOI: 10.1016/j.omtn.2024.102278
Evaluation of the effect of RNA secondary structure on Cas13d-mediated target RNA cleavage
Abstract
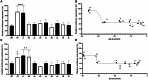
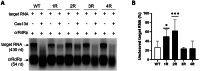
The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas13d system was adapted as a powerful tool for targeting viral RNA sequences, making it a promising approach for antiviral strategies. Understanding the influence of template RNA structure on Cas13d binding and cleavage efficiency is crucial for optimizing its therapeutic potential. In this study, we investigated the effect of local RNA secondary structure on Cas13d activity. To do so, we varied the stability of a hairpin structure containing the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) target sequence, allowing us to determine the threshold RNA stability at which Cas13d activity is affected. Our results demonstrate that Cas13d possesses the ability to effectively bind and cleave highly stable RNA structures. Notably, we only observed a decrease in Cas13d activity in the case of exceptionally stable RNA hairpins with completely base-paired stems, which are rarely encountered in natural RNA molecules. A comparison of Cas13d and RNA interference (RNAi)-mediated cleavage of the same RNA targets demonstrated that RNAi is more sensitive for local target RNA structures than Cas13d. These results underscore the suitability of the CRISPR-Cas13d system for targeting viruses with highly structured RNA genomes.
Keywords: CRISPR-Cas13d; MT: RNA/DNA Editing; RNA interference; RNA structure; SARS-CoV-2; cleavage activity; gene editing; targeting efficiency.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

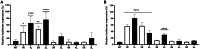
Similar articles
-
Cas13d: A New Molecular Scissor for Transcriptome Engineering.Front Cell Dev Biol. 2022 Mar 31;10:866800. doi: 10.3389/fcell.2022.866800. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35433685 Free PMC article. Review.
-
Massively parallel profiling of RNA-targeting CRISPR-Cas13d.Nat Commun. 2024 Jan 12;15(1):498. doi: 10.1038/s41467-024-44738-w. Nat Commun. 2024. PMID: 38216559 Free PMC article.
-
Fighting RNA viruses with a gold nanoparticle Cas13d gene-editing armor.Mol Ther Nucleic Acids. 2025 Apr 17;36(2):102540. doi: 10.1016/j.omtn.2025.102540. eCollection 2025 Jun 10. Mol Ther Nucleic Acids. 2025. PMID: 40391300 Free PMC article.
-
Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs.Genome Biol. 2021 Jan 21;22(1):41. doi: 10.1186/s13059-021-02263-9. Genome Biol. 2021. PMID: 33478577 Free PMC article.
-
Manipulation of genes could inhibit SARS-CoV-2 infection that causes COVID-19 pandemics.Exp Biol Med (Maywood). 2021 Jul;246(14):1643-1649. doi: 10.1177/15353702211008106. Epub 2021 Apr 25. Exp Biol Med (Maywood). 2021. PMID: 33899542 Free PMC article. Review.
Cited by
-
CRISPR-Cas13d: RNA's own Jedi Master in the fight against viral darkness.Mol Ther Nucleic Acids. 2024 Sep 20;35(4):102321. doi: 10.1016/j.omtn.2024.102321. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39380713 Free PMC article. No abstract available.
-
Current updates on the structural and functional aspects of the CRISPR/Cas13 system for RNA targeting and editing: A next‑generation tool for cancer management (Review).Int J Oncol. 2025 May;66(5):42. doi: 10.3892/ijo.2025.5748. Epub 2025 May 9. Int J Oncol. 2025. PMID: 40342053 Free PMC article. Review.
References
-
- Mojica F.J.M., Juez G., Rodriguezvalera F. Transcription at Different Salinities of Haloferax-Mediterranei Sequences Adjacent to Partially Modified Psti Sites. Mol. Microbiol. 1993;9:613–621. - PubMed
-
- Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

