Circulation of new lineages of RSV-A and RSV-B in Kuwait shows high diversity in the N- and O-linked glycosylation sites in the G protein between 2020 and 2022
- PMID: 39220282
- PMCID: PMC11362131
- DOI: 10.3389/fcimb.2024.1445115
Circulation of new lineages of RSV-A and RSV-B in Kuwait shows high diversity in the N- and O-linked glycosylation sites in the G protein between 2020 and 2022
Abstract
The human respiratory syncytial virus (RSV) is a significant health concern, particularly for infants, young children, and the elderly. This virus is known to evolve continuously due to environmental factors and herd immunity. In light of this, our study aimed to analyze the genetic variability of the G protein in RSV-A and RSV-B genotypes in Kuwait from 2020 to 2022. Between January 2020 and September 2022, we collected 490 respiratory samples from hospitalized patients with acute respiratory tract infections. These samples were tested and confirmed positive for RSV using multiplex Real-Time PCR. Subsequently, the samples underwent nucleic acid sequencing using the advanced Nanopore sequencing technology to analyze the full-length G gene. Sequence analysis showed that 64 isolates (76%) were RSV-A, and 20 isolates (24%) were RSV-B. The G genes of RSV-A belonged to genotype GA2.3.5, while all the RSV-B genotypes belonged to GB5.0.5a. New lineages and sub-lineages of RSV-A and RSV-B were detected, indicating the circulation of new strains in Kuwait. Many unique and new amino acid changes, including insertions, were found in the G proteins of Kuwaiti isolates, with the highest variability in the second hypervariable region. An increased number of N and O-linked glycosylation sites were also identified in the G protein, which could speculate to alter the antigenicity of RSV. The identified changes in the G protein of RSV-A and RSV-B genotypes might result from immune pressure and could affect the antigenic characteristics of circulating strains in Kuwait. This could potentially lead to new RSV variants that can evade the immune response. Our in-depth analysis of the G proteins of both RSV-A and RSV-B could aid in the development of more potent treatments and vaccines.
Keywords: G protein; Kuwait; glycosylation; nanopore sequencing; respiratory syncytial virus.
Copyright © 2024 Madi, Sadeq, Safar, Al-Adwani and Al-Turab.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
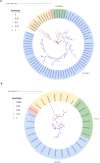

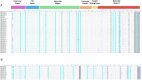


References
-
- Ahmed A., Haider S. H., Parveen S., Arshad M., Alsenaidy H. A., Baaboud A. O., et al. (2016). Co-circulation of 72bp duplication group A and 60bp duplication group B respiratory syncytial virus (RSV) strains in Riyadh, Saudi Arabia during 2014. PLoS One. 11, e0166145. doi: 10.1371/journal.pone.0166145 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

