Synthesis, Structural Characterization, and Computational Studies of Novel Co(II) and Zn(II) Fluoroquinoline Complexes for Antibacterial and Antioxidant Activities
- PMID: 39220483
- PMCID: PMC11359626
- DOI: 10.1021/acsomega.4c05560
Synthesis, Structural Characterization, and Computational Studies of Novel Co(II) and Zn(II) Fluoroquinoline Complexes for Antibacterial and Antioxidant Activities
Abstract
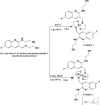
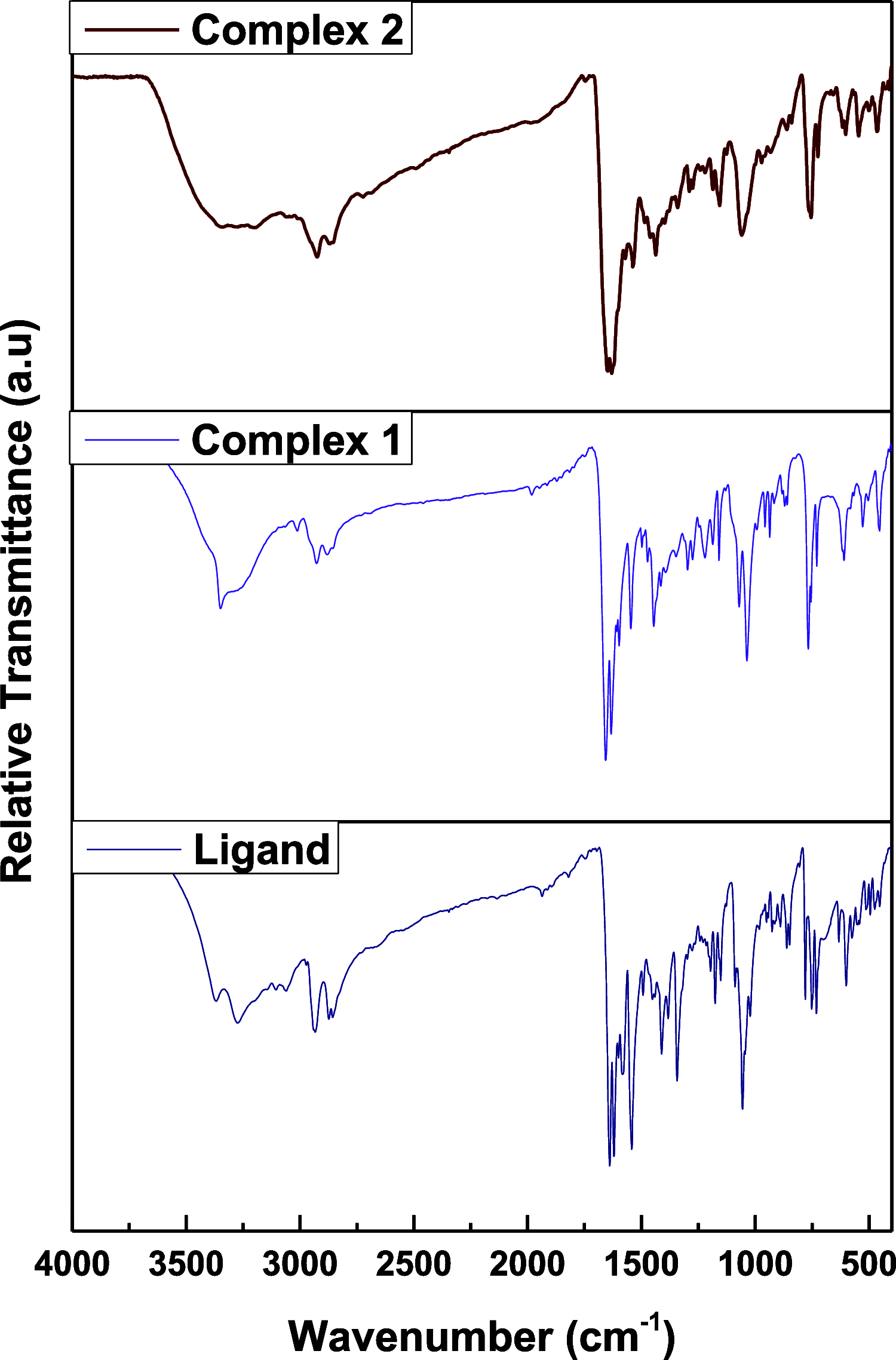
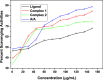
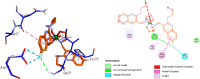
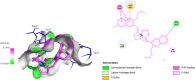
Research into heterocyclic ligands has increased in popularity due to their versatile applications in the biomedical field. Quinoline derivatives with their transition metal complexes are popular scaffolding molecules in the ongoing pursuit of newer and more effective bioactive molecules. Subsequently, this work reports on the synthesis and possible biological application of new Zn(II) and Co(II) complexes with a bidentate quinoline derivative ligand (H2 L), [(H2 L):(E)-2-(((6-fluoro-2-((2-hydroxyethyl)amino)quinolin-3-yl)methylene)amino)ethanol]. The ligand and its metal complexes were structurally characterized by spectroscopic methods (1H NMR, 13C NMR, Fourier transform infrared (FTIR), UV-vis, fluorescence, and mass spectroscopy), as well as by thermogravimetric and elemental analysis methods. The spectroscopic findings were further supported by density functional theory (DFT) and time-dependent (TD)-DFT calculations. The biological application was examined by investigating the inhibitory action of the complexes against bacterial strains using diffusion and agar dilution methods, and their profiles against two Gram-positive and Gram-negative bacterial strains were supported by molecular docking analysis. To rationalize the in vitro activity and establish the possible mechanism of action, the interactions and binding affinity of the ligand and complexes were investigated against three different bacterial enzymes (Escherichia coli DNA gyrase (PDB ID 6f86), E. coli dihydrofolate reductase B (PDB ID: 7r6g), and Staphylococcus aureus tyrosyl-tRNA synthetase (PDB ID: 1JIJ)) using AutoDock with the standard protocol. The MIC value of 0.20 μg/mL for zinc complex against E. coli and associated binding affinities -7.2 and -9.9 kcal/mol with DNA gyrase (PDB ID 6f86) and dihydrofolate reductase B (PDB ID: 7r6g), as well as the MIC value of 2.4 μg/mL for cobalt(II) complex against Staphylococcus aureus and the associated binding affinity of -10.5 kcal/mol with tyrosyl-tRNA synthetase (PDB ID: 1JIJ), revealed that the complexes' inhibitory actions were strong and comparable with those of the standard drug in the experiments. In addition, the ability of the new quinoline-based complexes to scavenge 1,1-diphenyl-picrylhydrazyl radicals was investigated; the findings suggested that the complexes exhibit potent antioxidant activities, which may be of therapeutic significance.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
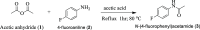

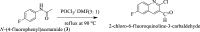











References
-
- Pinz M. P.; Reis A. S.; de Oliveira R. L.; Voss G. T.; Vogt A. G.; Sacramento M. do.; Roehrs J. A.; Alves D.; Luchese C.; Wilhelm E. A. 7-Chloro-4-Phenylsulfonyl Quinoline, a New Antinociceptive and Anti-Inflammatory Molecule: Structural Improvement of a Quinoline Derivate with Pharmacological Activity. Regul. Toxicol. Pharmacol. 2017, 90, 72–77. 10.1016/j.yrtph.2017.08.014. - DOI - PubMed
-
- Zeleke D.; Eswaramoorthy R.; Belay Z.; Melaku Y. Synthesis and Antibacterial, Antioxidant, and Molecular Docking Analysis of Some Novel Quinoline Derivatives. J. Chem. 2020, 2020, 1–16. 10.1155/2020/1324096. - DOI
-
- Murugavel S.; Jacob Prasanna Stephen C. S.; Subashini R.; AnanthaKrishnan D. Synthesis, Structural Elucidation, Antioxidant, CT-DNA Binding and Molecular Docking Studies of Novel Chloroquinoline Derivatives: Promising Antioxidant and Anti-Diabetic Agents. J. Photochem. Photobiol., B 2017, 173 (March), 216–230. 10.1016/j.jphotobiol.2017.05.043. - DOI - PubMed
LinkOut - more resources
Full Text Sources
