Epi-Cyclophellitol Cyclosulfate, a Mechanism-Based Endoplasmic Reticulum α-Glucosidase II Inhibitor, Blocks Replication of SARS-CoV-2 and Other Coronaviruses
- PMID: 39220688
- PMCID: PMC11363342
- DOI: 10.1021/acscentsci.4c00506
Epi-Cyclophellitol Cyclosulfate, a Mechanism-Based Endoplasmic Reticulum α-Glucosidase II Inhibitor, Blocks Replication of SARS-CoV-2 and Other Coronaviruses
Abstract
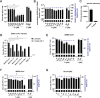
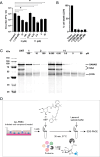
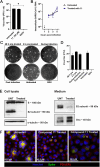
The combined inhibition of endoplasmic reticulum (ER) α-glucosidases I and II has been shown to inhibit replication of a broad range of viruses that rely on ER protein quality control. We found, by screening a panel of deoxynojirimycin and cyclitol glycomimetics, that the mechanism-based ER α-glucosidase II inhibitor, 1,6-epi-cyclophellitol cyclosulfate, potently blocks SARS-CoV-2 replication in lung epithelial cells, halting intracellular generation of mature spike protein, reducing production of infectious progeny, and leading to reduced syncytium formation. Through activity-based protein profiling, we confirmed ER α-glucosidase II inhibition in primary airway epithelial cells, grown at the air-liquid interface. 1,6-epi-Cyclophellitol cyclosulfate inhibits early pandemic and more recent SARS-CoV-2 variants, as well as SARS-CoV and MERS-CoV. The reported antiviral activity is comparable to the best-in-class described glucosidase inhibitors, all competitive inhibitors also targeting ER α-glucosidase I and other glycoprocessing enzymes not involved in ER protein quality control. We propose selective blocking ER-resident α-glucosidase II in a covalent and irreversible manner as a new strategy in the search for effective antiviral agents targeting SARS-CoV-2 and other viruses that rely on ER protein quality control.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
