From molecular subgroups to molecular targeted therapy in rheumatoid arthritis: A bioinformatics approach
- PMID: 39220908
- PMCID: PMC11365346
- DOI: 10.1016/j.heliyon.2024.e35774
From molecular subgroups to molecular targeted therapy in rheumatoid arthritis: A bioinformatics approach
Abstract
1background: Rheumatoid Arthritis (RA) is a heterogeneous autoimmune disease with multiple unidentified pathogenic factors. The inconsistency between molecular subgroups poses challenges for early diagnosis and personalized treatment strategies. In this study, we aimed to accurately distinguish RA patients at the transcriptome level using bioinformatics methods.
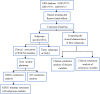
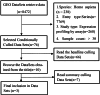
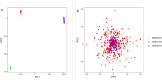
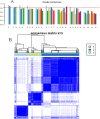
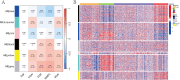
2methods: We collected a total of 362 transcriptome datasets from RA patients in three independent samples from the GEO database. Consensus clustering was performed to identify molecular subgroups, and clinical features were assessed. Differential analysis was employed to annotate the biological functions of specifically upregulated genes between subgroups.
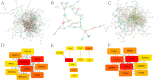
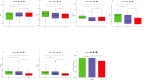
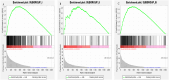
3results: Based on consensus clustering of RA samples, we identified three robust molecular subgroups, with Subgroup III representing the high-risk subgroup and Subgroup II exhibiting a milder phenotype, possibly associated with relatively higher levels of autophagic ability. Subgroup I showed biological functions mainly related to viral infections, cellular metabolism, protein synthesis, and inflammatory responses. Subgroup II involved autophagy of mitochondria and organelles, protein localization, and organelle disassembly pathways, suggesting heterogeneity in the autophagy process of mitochondria that may play a protective role in inflammatory diseases. Subgroup III represented a high-risk subgroup with pathological processes including abnormal amyloid precursor protein activation, promotion of inflammatory response, and cell proliferation.
4conclusion: The classification of the RA dataset revealed pathological heterogeneity among different subgroups, providing new insights and a basis for understanding the molecular mechanisms of RA, identifying potential therapeutic targets, and developing personalized treatment approaches.
Keywords: Diagnosis; Heterogeneity; Molecular subgroup; Rheumatoid arthritis; Transcriptomics.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
LinkOut - more resources
Full Text Sources

