TCR-H: explainable machine learning prediction of T-cell receptor epitope binding on unseen datasets
- PMID: 39221256
- PMCID: PMC11361934
- DOI: 10.3389/fimmu.2024.1426173
TCR-H: explainable machine learning prediction of T-cell receptor epitope binding on unseen datasets
Abstract
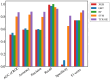
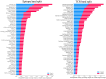
Artificial-intelligence and machine-learning (AI/ML) approaches to predicting T-cell receptor (TCR)-epitope specificity achieve high performance metrics on test datasets which include sequences that are also part of the training set but fail to generalize to test sets consisting of epitopes and TCRs that are absent from the training set, i.e., are 'unseen' during training of the ML model. We present TCR-H, a supervised classification Support Vector Machines model using physicochemical features trained on the largest dataset available to date using only experimentally validated non-binders as negative datapoints. TCR-H exhibits an area under the curve of the receiver-operator characteristic (AUC of ROC) of 0.87 for epitope 'hard splitting' (i.e., on test sets with all epitopes unseen during ML training), 0.92 for TCR hard splitting and 0.89 for 'strict splitting' in which neither the epitopes nor the TCRs in the test set are seen in the training data. Furthermore, we employ the SHAP (Shapley additive explanations) eXplainable AI (XAI) method for post hoc interrogation to interpret the models trained with different hard splits, shedding light on the key physiochemical features driving model predictions. TCR-H thus represents a significant step towards general applicability and explainability of epitope:TCR specificity prediction.
Keywords: T-cell receptor; adaptive immunity T-cell receptor; antigen; epitope; explainable machine learning; machine learning; physicochemical features; physicochemical model.
Copyright © 2024 T., Demerdash and Smith.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

