How does CHD4 slide nucleosomes?
- PMID: 39221830
- PMCID: PMC11555702
- DOI: 10.1042/BST20230070
How does CHD4 slide nucleosomes?
Abstract
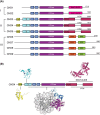
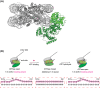
Chromatin remodelling enzymes reposition nucleosomes throughout the genome to regulate the rate of transcription and other processes. These enzymes have been studied intensively since the 1990s, and yet the mechanism by which they operate has only very recently come into focus, following advances in cryoelectron microscopy and single-molecule biophysics. CHD4 is an essential and ubiquitous chromatin remodelling enzyme that until recently has received less attention than remodellers such as Snf2 and CHD1. Here we review what recent work in the field has taught us about how CHD4 reshapes the genome. Cryoelectron microscopy and single-molecule studies demonstrate that CHD4 shares a central remodelling mechanism with most other chromatin remodellers. At the same time, differences between CHD4 and other chromatin remodellers result from the actions of auxiliary domains that regulate remodeller activity by for example: (1) making differential interactions with nucleosomal epitopes such as the acidic patch and the N-terminal tail of histone H4, and (2) inducing the formation of distinct multi-protein remodelling complexes (e.g. NuRD vs ChAHP). Thus, although we have learned much about remodeller activity, there is still clearly much more waiting to be revealed.
Keywords: CHD4; chromatin; enzyme activity.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

