Tryptophan 2,3-dioxygenase-positive matrix fibroblasts fuel breast cancer lung metastasis via kynurenine-mediated ferroptosis resistance of metastatic cells and T cell dysfunction
- PMID: 39221971
- PMCID: PMC11570772
- DOI: 10.1002/cac2.12608
Tryptophan 2,3-dioxygenase-positive matrix fibroblasts fuel breast cancer lung metastasis via kynurenine-mediated ferroptosis resistance of metastatic cells and T cell dysfunction
Abstract
Background: Tumor metastasis is a major threat to cancer patient survival. The organ-specific niche plays a pivotal role in tumor organotropic metastasis. Fibroblasts serve as a vital component of the metastatic microenvironment, but how heterogeneous metastasis-associated fibroblasts (MAFs) promote organotropic metastasis is poorly characterized. Here, we aimed to decipher the heterogeneity of MAFs and elucidate the distinct roles of these fibroblasts in pulmonary metastasis formation in breast cancer.
Methods: Mouse models of breast cancer pulmonary metastasis were established using an in vivo selection method of repeated injections of metastatic cells purified from the mouse lung. Single-cell RNA-sequencing (scRNA-seq) was employed to investigate the heterogeneity of MAFs. Transgenic mice were used to examine the contribution of tryptophan 2,3-dioxygenase-positive matrix fibroblasts (TDO2+ MFs) in lung metastasis.
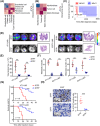
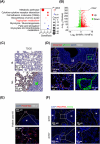
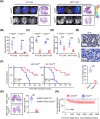
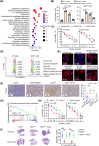
Results: We uncovered 3 subtypes of MAFs in the lung metastatic microenvironment, and their transcriptome profiles changed dynamically as lung metastasis evolved. As the predominant subtype, MFs were exclusively marked by platelet-derived growth factor receptor alpha (PDGFRA) and mainly located on the edge of the metastasis, and T cells were enriched around MFs. Notably, high MF signatures were significantly associated with poor survival in breast cancer patients. Lung metastases were markedly diminished, and the suppression of T cells was dramatically attenuated in MF-depleted experimental metastatic mouse models. We found that TDO2+ MFs controlled pulmonary metastasis by producing kynurenine (KYN), which upregulated ferritin heavy chain 1 (FTH1) level in disseminated tumor cells (DTCs), enabling DTCs to resist ferroptosis. Moreover, TDO2+ MF-secreted chemokines C-C motif chemokine ligand 8 (CCL8) and C-C motif chemokine ligand 11 (CCL11) recruited T cells. TDO2+ MF-derived KYN induced T cell dysfunction. Conditional knockout of Tdo2 in MFs diminished lung metastasis and enhanced immune activation.
Conclusions: Our study reveals crucial roles of TDO2+ MFs in promoting lung metastasis and DTCs' immune evasion in the metastatic niche. It suggests that targeting the metabolism of lung-specific stromal cells may be an effective treatment strategy for breast cancer patients with lung metastasis.
Keywords: Ferroptosis; Lung metastasis; Matrix fibroblasts; T cell dysfunction; Tryptophan 2,3‐dioxygenase.
© 2024 The Author(s). Cancer Communications published by John Wiley & Sons Australia, Ltd on behalf of Sun Yat‐sen University Cancer Center.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Gao Y, Wang Y, He B, Pan Y, Zhou D, Xiong M, et al. An Enzyme‐Loaded Metal‐Organic Framework‐Assisted Microfluidic Platform Enables Single‐Cell Metabolite Analysis. Angew Chem Int Ed Engl. 2023;62(31):e202302000. - PubMed
MeSH terms
Substances
Grants and funding
- BJRC202025/the Outstanding Postgraduate Fund of Chongqing Medical University
- R21 CA280458/CA/NCI NIH HHS/United States
- BJRC202021/the Outstanding Postgraduate Fund of Chongqing Medical University
- R01 CA151610/CA/NCI NIH HHS/United States
- NSFC81874199/National Natural Science Foundation of China
- CYB22218/the Chongqing Graduate Research and Innovation Project of the Chongqing Education Committee
- MOST2018YFE0113700/National Key Projects of Ministry of Science and Technology of China
- NSFC82173155/National Natural Science Foundation of China
- 2019-R10005/the Outstanding Professorship Program of Chongqing Medical University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

