Genome-wide CRISPR/Cas9 library screen identifies C16orf62 as a host dependency factor for porcine deltacoronavirus infection
- PMID: 39222358
- PMCID: PMC11404382
- DOI: 10.1080/22221751.2024.2400559
Genome-wide CRISPR/Cas9 library screen identifies C16orf62 as a host dependency factor for porcine deltacoronavirus infection
Abstract
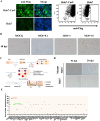
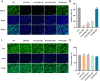
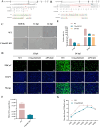
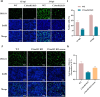
Porcine deltacoronavirus (PDCoV) is an emerging pathogen that can cause severe diarrhoea and high mortality in suckling piglets. Moreover, evidence of PDCoV infection in humans has raised concerns regarding potential public health risks. To identify potential therapeutic targets for PDCoV, we performed a genome-wide CRISPR/Cas9 library screening to find key host factors important to PDCoV infection. Several host genes in this screen were enriched, including ANPEP, which encodes the PDCoV receptor aminopeptidase N (APN). Furthermore, we discovered C16orf62, also known as the VPS35 endosomal protein sorting factor like (VPS35L), as an important host factor required for PDCoV infection. C16orf62 is an important component of the multiprotein retriever complex involved in protein recycling in the endosomal compartment and its gene knockout led to a remarkable decrease in the binding and internalization of PDCoV into host cells. While we did not find evidence for direct interaction between C16orf62 and the viral s (spike) protein, C16orf62 gene knockout was shown to downregulate APN expression at the cell surface. This study marks the first instance of a genome-wide CRISPR/Cas9-based screen tailored for PDCoV, revealing C16orf62 as a host factor required for PDCoV replication. These insights may provide promising avenues for the development of antiviral drugs against PDCoV infection.
Keywords: C16orf62; CRISPR/Cas9; Porcine deltacoronavirus; aminopeptidase N; host factor.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
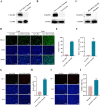
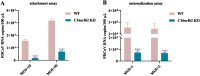
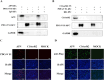
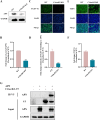
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
