Transforming kidney transplant monitoring with urine CXCL9 and CXCL10: practical clinical implementation
- PMID: 39223175
- PMCID: PMC11369285
- DOI: 10.1038/s41598-024-70390-x
Transforming kidney transplant monitoring with urine CXCL9 and CXCL10: practical clinical implementation
Abstract
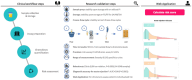
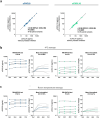
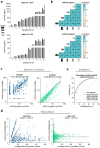
In kidney transplant recipients, urine CXCL9 and CXCL10 (uCXCL9/10) chemokines have reached a sufficiently high level of evidence to be recommended by the European Society of Organ Transplantation for the monitoring of immune quiescence. To assess the risk of acute rejection (AR), the advantage of uCXCL9/10 is their cost-effectiveness and their high diagnostic performance. Here, we evaluated the feasibility of a next-generation immunoassay for quantifying uCXCL9/10 levels. It demonstrated high efficiency with minimal workflow and a 90-min time to result. Preanalytical studies indicated stability of uCXCL9/10 levels and analytical studies confirmed excellent linearity and precision. In a cohort of 1048 samples collected at biopsy, the results correlated significantly with ELISA quantification and were integrated into a previously validated 8-parameter urine chemokine model. The next generation immunoassay achieved an accuracy of 0.84 for AR diagnosis. This study validates this technology as a robust, locally available and unexpensive platform and marks a significant step towards the widespread implementation of uCXCL9/10, for immune quiescence monitoring. Therefore, we developed an open-access web application using uCXCL9/10 to calculate AR risk and improve clinical decision-making to perform biopsy, ushering in a new era in kidney transplantation, where personalized, data-driven care becomes the norm.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

