Impact of KDM6B mosaic brain knockout on synaptic function and behavior
- PMID: 39223259
- PMCID: PMC11369245
- DOI: 10.1038/s41598-024-70728-5
Impact of KDM6B mosaic brain knockout on synaptic function and behavior
Abstract
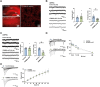
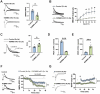
Autism spectrum disorders (ASD) are complex neurodevelopmental conditions characterized by impairments in social communication, repetitive behaviors, and restricted interests. Epigenetic modifications serve as critical regulators of gene expression playing a crucial role in controlling brain function and behavior. Lysine (K)-specific demethylase 6B (KDM6B), a stress-inducible H3K27me3 demethylase, has emerged as one of the highest ASD risk genes, but the precise effects of KDM6B mutations on neuronal activity and behavioral function remain elusive. Here we show the impact of KDM6B mosaic brain knockout on the manifestation of different autistic-like phenotypes including repetitive behaviors, social interaction, and significant cognitive deficits. Moreover, KDM6B mosaic knockout display abnormalities in hippocampal excitatory synaptic transmission decreasing NMDA receptor mediated synaptic transmission and plasticity. Understanding the intricate interplay between epigenetic modifications and neuronal function may provide novel insights into the pathophysiology of ASD and potentially inform the development of targeted therapeutic interventions.
Keywords: ASD; Behavior; Gene editing; KDM6B; NMDA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

