Retrotransposon addiction promotes centromere function via epigenetically activated small RNAs
- PMID: 39223305
- PMCID: PMC11410651
- DOI: 10.1038/s41477-024-01773-1
Retrotransposon addiction promotes centromere function via epigenetically activated small RNAs
Abstract
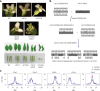
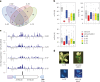
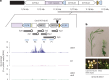
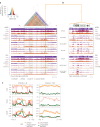
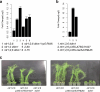
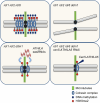
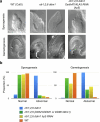
Retrotransposons have invaded eukaryotic centromeres in cycles of repeat expansion and purging, but the function of centromeric retrotransposons has remained unclear. In Arabidopsis, centromeric ATHILA retrotransposons give rise to epigenetically activated short interfering RNAs in mutants in DECREASE IN DNA METHYLATION1 (DDM1). Here we show that mutants that lose both DDM1 and RNA-dependent RNA polymerase have pleiotropic developmental defects and mis-segregate chromosome 5 during mitosis. Fertility and segregation defects are epigenetically inherited with centromere 5, and can be rescued by directing artificial small RNAs to ATHILA5 retrotransposons that interrupt tandem satellite repeats. Epigenetically activated short interfering RNAs promote pericentromeric condensation, chromosome cohesion and chromosome segregation in mitosis. We propose that insertion of ATHILA silences centromeric transcription, while simultaneously making centromere function dependent on retrotransposon small RNAs in the absence of DDM1. Parallels are made with the fission yeast Schizosaccharomyces pombe, where chromosome cohesion depends on RNA interference, and with humans, where chromosome segregation depends on both RNA interference and HELLSDDM1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Update of
-
Retrotransposon addiction promotes centromere function via epigenetically activated small RNAs.bioRxiv [Preprint]. 2023 Aug 3:2023.08.02.551486. doi: 10.1101/2023.08.02.551486. bioRxiv. 2023. Update in: Nat Plants. 2024 Sep;10(9):1304-1316. doi: 10.1038/s41477-024-01773-1. PMID: 37577592 Free PMC article. Updated. Preprint.
References
-
- Steiner, F. A. & Henikoff, S. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev.31, 28–35 (2015). - PubMed
-
- Presting, G. G. Centromeric retrotransposons and centromere function. Curr. Opin. Genet. Dev.49, 79–84 (2018). - PubMed
-
- Matzke, M. A. & Mosher, R. A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet.15, 394–408 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

