Targeting NLRP3 inhibits AML progression by inducing PERK/eIF2-mediated apoptosis
- PMID: 39223663
- PMCID: PMC11367831
- DOI: 10.1186/s12964-024-01777-6
Targeting NLRP3 inhibits AML progression by inducing PERK/eIF2-mediated apoptosis
Abstract
Background: Acute myeloid leukemia (AML) is characterized by the abnormal proliferation of myeloid precursor cells and presents significant challenges in treatment due to its heterogeneity. Recently, the NLRP3 inflammasome has emerged as a potential contributor to AML pathogenesis, although its precise mechanisms remain poorly understood.
Methods: Public genome datasets were utilized to evaluate the expression of NLRP3 inflammasome-related genes (IL-1β, IL-18, ASC, and NLRP3) in AML patients compared to healthy individuals. CRISPR/Cas9 technology was employed to generate NLRP3-deficient MOLM-13 AML cells, followed by comprehensive characterization using real-time PCR, western blotting, FACS analysis, and transmission electron and immunofluorescence microscopy. Proteomic analyses were conducted to identify NLRP3-dependent alterations in protein levels, with a focus on the eIF2 kinase PERK-mediated signaling pathways. Additionally, in vivo studies were performed using a leukemic mouse model to elucidate the pathogenic role of NLRP3 in AML.
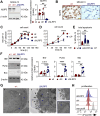
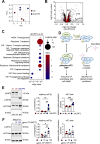
Results: Elevated expression of NLRP3 was significantly associated with diminished overall survival in AML patients. Genetic deletion, pharmacological inhibition and silencing by RNA interference of NLRP3 led to decreased AML cell survival through the induction of apoptosis. Proteomic analyses uncovered NLRP3-dependent alterations in protein translation, characterized by enhanced eIF2α phosphorylation in NLRP3-deficient AML cells. Moreover, inhibition of PERK-mediated eIF2α phosphorylation reduced apoptosis by downregulating pro-apoptotic Bcl-2 family members. In vivo studies demonstrated reduced leukemic burden in mice engrafted with NLRP3 knockout AML cells, as evidenced by alleviated leukemic symptoms.
Conclusion: Our findings elucidate the involvement of the NLRP3/PERK/eIF2 axis as a novel driver of AML cell survival. Targeting NLRP3-induced signaling pathways, particularly through the PERK/eIF2 axis, presents a promising therapeutic strategy for AML intervention. These insights into the role of the NLRP3 inflammasome offer potential avenues for improving the prognosis and treatment outcomes of AML patients.
Keywords: Acute myeloid leukemia; Apoptosis; PERK; eIF2α, NLRP3.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- 20102-F2001080-FPR/Cancer Cluster Salzburg
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- 20102-P1601064-FPR01-2017/Cancer Cluster Salzburg
- P33969/Austrian Science Fund
- P33969/Austrian Science Fund
- W1213/Austrian Science Fund
- P33969/Austrian Science Fund
- W1213/Austrian Science Fund
- W1213/Austrian Science Fund
- W1213/Austrian Science Fund
- W1213/Austrian Science Fund
- W1213/Austrian Science Fund
- W1213/Austrian Science Fund
- P33969/Austrian Science Fund
- ITAT1054/Interreg
- ITAT1054/Interreg
- ITAT1054/Interreg
- ITAT1054/Interreg
- ITAT1054/Interreg
- ITAT1054/Interreg
- 20102-F1901165-KZP/Salzburger Landesregierung
- 20102-F1901165-KZP/Salzburger Landesregierung
- 20102-F1901165-KZP/Salzburger Landesregierung
- DJCLS 04 R/2020/Jose Carreras
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

