Differential Pharmacodynamic Effects on Psoriatic Biomarkers by Guselkumab Versus Secukinumab Correlate with Long-Term Efficacy: An ECLIPSE Substudy
- PMID: 39224116
- PMCID: PMC11367549
- DOI: 10.1016/j.xjidi.2024.100297
Differential Pharmacodynamic Effects on Psoriatic Biomarkers by Guselkumab Versus Secukinumab Correlate with Long-Term Efficacy: An ECLIPSE Substudy
Abstract
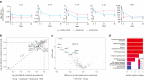
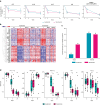
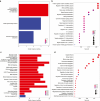
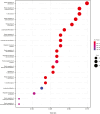
IL-23 is a cytokine produced by myeloid cells that drives the T helper 17 pathway and plays an essential role in the pathophysiology of plaque psoriasis. IL-23 activation initiates a cascade of cytokines subsequently inducing the expression of many psoriasis-related proteins. This study aimed to better understand the underlying mechanisms driving the differences between IL-23 and IL-17A blockade in patients with psoriasis and their implications for durability of clinical responses. Serum and/or skin biopsies were isolated from patients treated with guselkumab or secukinumab for evaluation of potential biomarkers of pharmacodynamic response to treatment. Guselkumab treatment led to significantly greater reductions of IL-17F and IL-22 serum levels than treatment with secukinumab at weeks 24 and 48, demonstrating sustained regulation of the IL-23/T helper 17 pathway. Analyses of proteomic and transcriptomic profiles of patient sera and skin biopsies demonstrated differential regulation of proteins involved in chemokine, TNF, and relevant immune signaling pathways to a greater degree with guselkumab than with secukinumab treatment. These data provide insights into the differences between the mechanisms and impact of IL-23 and IL-17A blockade in psoriasis, with implications for efficacy observations and treatment paradigms. Trial Registration: The original study was registered at ClinicalTrials.gov (NCT03090100).
Keywords: Guselkumab; PD; Psoriasis; Secukinumab.
© 2024 The Authors.
Figures





References
-
- Blauvelt A., Ferris L.K., Yamauchi P.S., Qureshi A., Leonardi C.L., Farahi K., et al. Extension of ustekinumab maintenance dosing interval in moderate-to-severe psoriasis: results of a phase IIIb, randomized, double-blinded, active-controlled, multicentre study (PSTELLAR) Br J Dermatol. 2017;177:1552–1561. - PubMed
-
- Blauvelt A., Papp K.A., Griffiths C.E., Randazzo B., Wasfi Y., Shen Y.K., et al. Efficacy and safety of guselkumab, an anti-interleukin-23 monoclonal antibody, compared with adalimumab for the continuous treatment of patients with moderate to severe psoriasis: results from the phase III, double-blinded, placebo- and active comparator-controlled VOYAGE 1 trial. J Am Acad Dermatol. 2017;76:405–417. - PubMed
-
- Blumberg H., Conklin D., Xu W.F., Grossmann A., Brender T., Carollo S., et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Medical

