Functional genomics identifies a small secreted protein that plays a role during the biotrophic to necrotrophic shift in the root rot pathogen Phytophthora medicaginis
- PMID: 39224851
- PMCID: PMC11366588
- DOI: 10.3389/fpls.2024.1439020
Functional genomics identifies a small secreted protein that plays a role during the biotrophic to necrotrophic shift in the root rot pathogen Phytophthora medicaginis
Abstract
Introduction: Hemibiotrophic Phytophthora are a group of agriculturally and ecologically important pathogenic oomycetes causing severe decline in plant growth and fitness. The lifestyle of these pathogens consists of an initial biotrophic phase followed by a switch to a necrotrophic phase in the latter stages of infection. Between these two phases is the biotrophic to necrotrophic switch (BNS) phase, the timing and controls of which are not well understood particularly in Phytophthora spp. where host resistance has a purely quantitative genetic basis.
Methods: To investigate this we sequenced and annotated the genome of Phytophthora medicaginis, causal agent of root rot and substantial yield losses to Fabaceae hosts. We analyzed the transcriptome of P. medicaginis across three phases of colonization of a susceptible chickpea host (Cicer arietinum) and performed co-regulatory analysis to identify putative small secreted protein (SSP) effectors that influence timing of the BNS in a quantitative pathosystem.
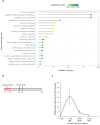
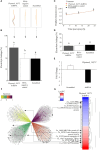
Results: The genome of P. medicaginis is ~78 Mb, comparable to P. fragariae and P. rubi which also cause root rot. Despite this, it encodes the second smallest number of RxLR (arginine-any amino acid-leucine-arginine) containing proteins of currently sequenced Phytophthora species. Only quantitative resistance is known in chickpea to P. medicaginis, however, we found that many RxLR, Crinkler (CRN), and Nep1-like protein (NLP) proteins and carbohydrate active enzymes (CAZymes) were regulated during infection. Characterization of one of these, Phytmed_10271, which encodes an RxLR effector demonstrates that it plays a role in the timing of the BNS phase and root cell death.
Discussion: These findings provide an important framework and resource for understanding the role of pathogenicity factors in purely quantitative Phytophthora pathosystems and their implications to the timing of the BNS phase.
Keywords: co-expression network; effector; genome; hemibiotrophic pathogenesis; immune manipulation.
Copyright © 2024 Coles, Bithell, Jeffries, Cuddy and Plett.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adams T. M., Armitage A. D., Sobczyk M. K., Bates H. J., Tabima J. F., Kronmiller B. A., et al. . (2020). Genomic investigation of the strawberry pathogen Phytophthora fragariae indicates pathogenicity is associated with transcriptional variation in three key races. Front. Microbiol. 11, 490. doi: 10.3389/fmicb.2020.00490 - DOI - PMC - PubMed
-
- Ahumada R., Rotella A., Slippers B., Wingfield M. J. (2013). Pathogenicity and sporulation of Phytophthora pinifolia on Pinus radiata in Chile. Australas. Plant Pathol. 42, 413–420. doi: 10.1007/s13313-013-0212-4 - DOI
-
- Alexa A., Rahnenführer J. (2009). Gene set enrichment analysis with topGO. Bioconduct. Improv. 27, 1–26.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

