Somatic RAP1B gain-of-function variant underlies isolated thrombocytopenia and immunodeficiency
- PMID: 39225097
- PMCID: PMC11364392
- DOI: 10.1172/JCI169994
Somatic RAP1B gain-of-function variant underlies isolated thrombocytopenia and immunodeficiency
Abstract
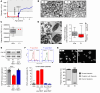
The ubiquitously expressed small GTPase Ras-related protein 1B (RAP1B) acts as a molecular switch that regulates cell signaling, cytoskeletal remodeling, and cell trafficking and activates integrins in platelets and lymphocytes. The residue G12 in the P-loop is required for the RAP1B-GTPase conformational switch. Heterozygous germline RAP1B variants have been described in patients with syndromic thrombocytopenia. However, the causality and pathophysiological impact remained unexplored. We report a boy with neonatal thrombocytopenia, combined immunodeficiency, neutropenia, and monocytopenia caused by a heterozygous de novo single nucleotide substitution, c.35G>A (p.G12E) in RAP1B. We demonstrate that G12E and the previously described G12V and G60R were gain-of-function variants that increased RAP1B activation, talin recruitment, and integrin activation, thereby modifying late responses such as platelet activation, T cell proliferation, and migration. We show that in our patient, G12E was a somatic variant whose allele frequency decreased over time in the peripheral immune compartment, but remained stable in bone marrow cells, suggesting a differential effect in distinct cell populations. Allogeneic hematopoietic stem cell transplantation fully restored the patient's hemato-immunological phenotype. Our findings define monoallelic RAP1B gain-of-function variants as a cause for constitutive immunodeficiency and thrombocytopenia. The phenotypic spectrum ranged from isolated hematological manifestations in our patient with somatic mosaicism to complex syndromic features in patients with reported germline RAP1B variants.
Keywords: Cell migration/adhesion; Genetic diseases; Hematology; Immunology; Platelets.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

