Novel Arf1 Inhibitors Drive Cancer Stem Cell Aging and Potentiate Anti-Tumor Immunity
- PMID: 39225354
- PMCID: PMC11497069
- DOI: 10.1002/advs.202404442
Novel Arf1 Inhibitors Drive Cancer Stem Cell Aging and Potentiate Anti-Tumor Immunity
Abstract
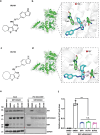
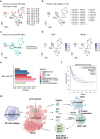
The small G protein Arf1 has been identified as playing a selective role in supporting cancer stem cells (CSCs), making it an attractive target for cancer therapy. However, the current Arf1 inhibitors have limited translational potential due to their high toxicity and low specificity. In this study, two new potent small-molecule inhibitors of Arf1, identified as DU101 and DU102, for cancer therapy are introduced. Preclinical tumor models demonstrate that these inhibitors triggered a cascade of aging in CSCs and enhance anti-tumor immunity in mouse cancer and PDX models. Through single-cell sequencing, the remodeling of the tumor immune microenvironment induced by these new Arf1 inhibitors is analyzed and an increase in tumor-associated CD8+ CD4+ double-positive T (DPT) cells is identified. These DPT cells exhibit superior features of active CD8 single-positive T cells and a higher percentage of TCF1+PD-1+, characteristic of stem-like T cells. The frequency of tumor-infiltrating stem-like DPT cells correlates with better disease-free survival (DFS) in cancer patients, indicating that these inhibitors may offer a novel cancer immunotherapy strategy by converting the cold tumor immune microenvironment into a hot one, thus expanding the potential for immunotherapy in cancer patients.
Keywords: Arf1 inhibitor; anti‐tumor immunity; cancer stem cell; superior T cells; trans‐cellular signaling.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
