Integrated epigenomic exposure signature discovery
- PMID: 39225561
- PMCID: PMC11404615
- DOI: 10.1080/17501911.2024.2375187
Integrated epigenomic exposure signature discovery
Abstract
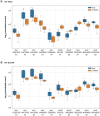
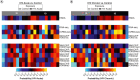
Aim: The epigenome influences gene regulation and phenotypes in response to exposures. Epigenome assessment can determine exposure history aiding in diagnosis.Materials & methods: Here we developed and implemented a machine learning algorithm, the exposure signature discovery algorithm (ESDA), to identify the most important features present in multiple epigenomic and transcriptomic datasets to produce an integrated exposure signature (ES).Results: Signatures were developed for seven exposures including Staphylococcus aureus, human immunodeficiency virus, SARS-CoV-2, influenza A (H3N2) virus and Bacillus anthracis vaccinations. ESs differed in the assays and features selected and predictive value.Conclusion: Integrated ESs can potentially be utilized for diagnosis or forensic attribution. The ESDA identifies the most distinguishing features enabling diagnostic panel development for future precision health deployment.
Keywords: diagnostics; epigenomics; exposure health; infection; machine learning; multi-omics; transcriptomics.
Plain language summary
This article introduces ESDA, a new analytic tool for integrating multiple data types to identify the most distinguishing features following an exposure. Using the ESDA, we were able to identify signatures of infectious diseases. The results of the study indicate that integration of multiple types of large datasets can be used to identify distinguishing features for infectious diseases. Understanding the changes from different exposures will enable development of diagnostic tests for infectious diseases that target responses from the patient. Using the ESDA, we will be able to build a database of human response signatures to different infections and simplify diagnostic testing in the future.
Conflict of interest statement
Barinthus Biotherapeutics is a company with financial interest in the influenza vaccine. The work here utilizes samples from their clinical trial provided by TGE and FC. The authors have no other competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript apart from those disclosed.
Figures




References
-
- Chakraborty R, Burns B. Systemic Inflammatory Response Syndrome. Treasure Island (FL): StatPearls Publishing; 2023. - PubMed
-
- Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci. 2018;115(52):E12353–E12362. doi: 10.1073/pnas.1809700115 - DOI - PMC - PubMed
-
• Demonstrates the diagnostic power of the host response to enable healthcare providers to better determine cause of respiratory infections and treat patients accordingly.
-
- Evans TG, Bussey L, Eagling-Vose E, et al. Efficacy and safety of a universal influenza A vaccine (MVA-NP+M1) in adults when given after seasonal quadrivalent influenza vaccine immunisation (FLU009): a phase 2b, randomised, double-blind trial. Lancet Infect Dis. 2022;22(6):857–866. doi: 10.1016/S1473-3099(21)00702-7 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
