Platelet-Derived Growth Factor C Facilitates Malignant Behavior of Pancreatic Ductal Adenocarcinoma by Regulating SREBP1 Mediated Lipid Metabolism
- PMID: 39225567
- PMCID: PMC11516052
- DOI: 10.1002/advs.202407069
Platelet-Derived Growth Factor C Facilitates Malignant Behavior of Pancreatic Ductal Adenocarcinoma by Regulating SREBP1 Mediated Lipid Metabolism
Abstract
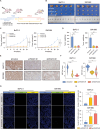
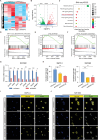
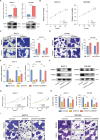
Lipid metabolism reprogramming stands as a fundamental hallmark of cancer cells. Unraveling the core regulators of lipid biosynthesis holds the potential to find promising therapeutic targets in pancreatic ductal adenocarcinoma (PDAC). Here, it is demonstrated that platelet-derived growth factor C (PDGFC) orchestrated lipid metabolism, thereby facilitated the malignant progression of PDAC. Expression of PDGFC is upregulated in PDAC cohorts and is corelated with a poor prognosis. Aberrantly high expression of PDGFC promoted proliferation and metastasis of PDAC both in vitro and in vivo. Mechanistically, PDGFC accelerated the malignant progression of PDAC by upregulating fatty acid accumulation through sterol regulatory element-binding protein 1 (SREBP1), a key transcription factor in lipid metabolism. Remarkably, Betulin, an inhibitor of SREBP1, demonstrated the capability to inhibit proliferation and metastasis of PDAC cell lines, along with attenuating the process of liver metastasis in vivo. Overall, the study underscores the pivotal role of PDGFC-mediated lipid metabolism in PDAC progression, suggesting PDGFC as a potential biomarker for PDAC metastasis. Targeting PDGFC-induced lipid metabolism emerges as a promising therapeutic strategy for metastatic PDAC, with the potential to improve clinical outcomes.
Keywords: PDAC; PDGFC; SREBP1; lipid metabolism; metastasis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C. D., Biankin A. V., Neale R. E., Tempero M., Tuveson D. A., Hruban R. H., Neoptolemos J. P., Nat. Rev. Dis. Primers 2016, 2, 16022. - PubMed
-
- Stathis A., Moore M. J., Nat. Rev. Clin. Oncol. 2010, 7, 163. - PubMed
-
- Lowery F. J., Yu D., Cancer Metastasis Rev. 2012, 31, 479. - PubMed
-
- Seruga B., Zhang H., Bernstein L. J., Tannock I. F., Nat. Rev. Cancer 2008, 8, 887. - PubMed
-
- Zou X., Tang X. Y., Qu Z. Y., Sun Z. W., Ji C. F., Li Y. J., Guo S. D., Int. J. Biol. Macromol. 2022, 202, 539. - PubMed
MeSH terms
Substances
Grants and funding
- 82072644/National Natural Science Foundation of China
- 82203473/National Natural Science Foundation of China
- 82203105/National Natural Science Foundation of China
- 82103401/National Natural Science Foundation of China
- 82103267/National Natural Science Foundation of China
- 2021A1515111104/Guangdong Basic and Applied Basic Research Foundation
- 2023A1515110914/Guangdong Basic and Applied Basic Research Foundation
- 2024A1515010487/Guangdong Basic and Applied Basic Research Foundation
- 2024A1515010400/Guangdong Basic and Applied Basic Research Foundation
- 2023M744050/China Postdoctoral Science Foundation
- GZC20233269/China Postdoctoral Science Foundation
- 2023A04J2211/Guangzhou Science and Technology Plan Project
- 2023P-ZD02/Guangzhou Clinical Major Technical Project Foundation
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
