Qki5 safeguards spinal motor neuron function by defining the motor neuron-specific transcriptome via pre-mRNA processing
- PMID: 39226364
- PMCID: PMC11406248
- DOI: 10.1073/pnas.2401531121
Qki5 safeguards spinal motor neuron function by defining the motor neuron-specific transcriptome via pre-mRNA processing
Abstract
Many RNA-binding proteins (RBPs) are linked to the dysregulation of RNA metabolism in motor neuron diseases (MNDs). However, the molecular mechanisms underlying MN vulnerability have yet to be elucidated. Here, we found that such an RBP, Quaking5 (Qki5), contributes to formation of the MN-specific transcriptome profile, termed "MN-ness," through the posttranscriptional network and maintenance of the mature MNs. Immunohistochemical analysis and single-cell RNA sequencing (scRNA-seq) revealed that Qki5 is predominantly expressed in MNs, but not in other neuronal populations of the spinal cord. Furthermore, comprehensive RNA sequencing (RNA-seq) analyses revealed that Qki5-dependent RNA regulation plays a pivotal role in generating the MN-specific transcriptome through pre-messenger ribonucleic acid (mRNA) splicing for the synapse-related molecules and c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) signaling pathways. Indeed, MN-specific ablation of the Qki5 caused neurodegeneration in postnatal mice and loss of Qki5 function resulted in the aberrant activation of stress-responsive JNK/SAPK pathway both in vitro and in vivo. These data suggested that Qki5 plays a crucial biological role in RNA regulation and safeguarding of MNs and might be associated with pathogenesis of MNDs.
Keywords: Quaking5; RNA-binding protein; alternative splicing; degeneration; motor neuron.
Conflict of interest statement
Competing interests statement:H.O. is a paid member of the Scientific Advisory Boards of San Bio Co., Ltd., and K Pharma Inc. M. Yano is a paid member of the Scientific Advisory Board of K Pharma Inc.
Figures
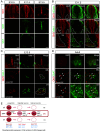



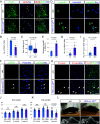
References
-
- Conn S. J., et al. , The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015). - PubMed
MeSH terms
Substances
Grants and funding
- JP20H00485a/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP19H03543/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP22H05589/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP23gm1710010/Japan Agency for Medical Research and Development (AMED)
- JP22K06879/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

