Decoding the pathogenesis of spermatogenic failure in cryptorchidism through single-cell transcriptomic profiling
- PMID: 39226895
- PMCID: PMC11528238
- DOI: 10.1016/j.xcrm.2024.101709
Decoding the pathogenesis of spermatogenic failure in cryptorchidism through single-cell transcriptomic profiling
Abstract
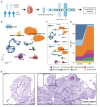
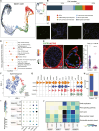
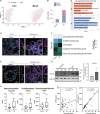
Cryptorchidism, commonly known as undescended testis, affects 1%-9% of male newborns, posing infertility and testis tumor risks. Despite its prevalence, the detailed pathophysiology underlying male infertility within cryptorchidism remains unclear. Here, we profile and analyze 46,644 single-cell transcriptomes from individual testicular cells obtained from adult males diagnosed with cryptorchidism and healthy controls. Spermatogenesis compromise in cryptorchidism links primarily to spermatogonium self-renewal and differentiation dysfunctions. We illuminate the involvement of testicular somatic cells, including immune cells, thereby unveiling the activation and degranulation of mast cells in cryptorchidism. Mast cells are identified as contributors to interstitial fibrosis via transforming growth factor β1 (TGF-β1) and cathepsin G secretion. Furthermore, significantly increased levels of secretory proteins indicate mast cell activation and testicular fibrosis in the seminal plasma of individuals with cryptorchidism compared to controls. These insights serve as valuable translational references, enriching our comprehension of testicular pathogenesis and informing more precise diagnosis and targeted therapeutic strategies for cryptorchidism.
Keywords: cryptorchidism; male infertility; spermatogonial stem cells; testicular interstitial fibrosis.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Florou M., Tsilidis K.K., Siomou E., Koletsa T., Syrnioti A., Spyridakis I., Kaselas C., Ntzani E.E. Orchidopexy for congenital cryptorchidism in childhood and adolescence and testicular cancer in adults: an updated systematic review and meta-analysis of observational studies. Eur. J. Pediatr. 2023;182:2499–2507. doi: 10.1007/s00431-023-04947-9. - DOI - PubMed
-
- van Brakel J., Kranse R., de Muinck Keizer-Schrama S.M.P.F., Hendriks A.E.J., de Jong F.H., Bangma C.H., Hazebroek F.W.J., Dohle G.R. Fertility potential in men with a history of congenital undescended testes: a long-term follow-up study. Andrology. 2013;1:100–108. doi: 10.1111/j.2047-2927.2012.00024.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

