Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch
- PMID: 39227374
- PMCID: PMC11372198
- DOI: 10.1038/s41467-024-51799-4
Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch
Abstract
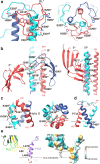
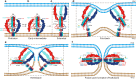
The fusion of viruses with cellular membranes is a critical step in the life cycle of enveloped viruses. This process is facilitated by viral fusion proteins, many of which are conformationally pH-sensitive. The specifics of how changes in pH initiate this fusion have remained largely elusive. This study presents the cryo-electron microscopy (cryo-EM) structures of a prototype class III fusion protein, GP64, in its prefusion and early intermediate states, revealing the structural intermediates accompanying the membrane fusion process. The structures identify the involvement of a pH-sensitive switch, comprising H23, H245, and H304, in sensing the low pH that triggers the initial step of membrane fusion. The pH sensing role of this switch is corroborated by assays of cell-cell syncytium formation and dual dye-labeling. The findings demonstrate that coordination between multiple histidine residues acts as a pH sensor and activator. The involvement of a multi-histidine switch in viral fusion is applicable to fusogens of human-infecting thogotoviruses and other viruses, which could lead to strategies for developing anti-viral therapies and vaccines.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 31971133/National Natural Science Foundation of China (National Science Foundation of China)
- 32172485/National Natural Science Foundation of China (National Science Foundation of China)
- 19PJ1407900/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 19JC1414000/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
- 22WZ2504100/Science and Technology Commission of Shanghai Municipality (Shanghai Municipal Science and Technology Commission)
LinkOut - more resources
Full Text Sources

