A molecular mechanism to diversify Ca2+ signaling downstream of Gs protein-coupled receptors
- PMID: 39227390
- PMCID: PMC11372221
- DOI: 10.1038/s41467-024-51991-6
A molecular mechanism to diversify Ca2+ signaling downstream of Gs protein-coupled receptors
Abstract
A long-held tenet in inositol-lipid signaling is that cleavage of membrane phosphoinositides by phospholipase Cβ (PLCβ) isozymes to increase cytosolic Ca2+ in living cells is exclusive to Gq- and Gi-sensitive G protein-coupled receptors (GPCRs). Here we extend this central tenet and show that Gs-GPCRs also partake in inositol-lipid signaling and thereby increase cytosolic Ca2+. By combining CRISPR/Cas9 genome editing to delete Gαs, the adenylyl cyclase isoforms 3 and 6, or the PLCβ1-4 isozymes, with pharmacological and genetic inhibition of Gq and G11, we pin down Gs-derived Gβγ as driver of a PLCβ2/3-mediated cytosolic Ca2+ release module. This module does not require but crosstalks with Gαs-dependent cAMP, demands Gαq to release PLCβ3 autoinhibition, but becomes Gq-independent with mutational disruption of the PLCβ3 autoinhibited state. Our findings uncover the key steps of a previously unappreciated mechanism utilized by mammalian cells to finetune their calcium signaling regulation through Gs-GPCRs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
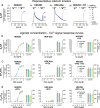
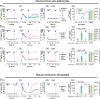
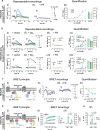


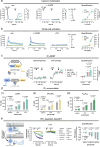


References
Publication types
MeSH terms
Substances
Grants and funding
- 214362475/GRK1873/2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 494832089 /GRK2873/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 290847012/FOR2372/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 214362475/GRK1873/2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 494832089 /GRK2873/Deutsche Forschungsgemeinschaft (German Research Foundation)
- JP21H04791/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP21H05113/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP21H05037/MEXT | Japan Society for the Promotion of Science (JSPS)
- JPJSBP120213501/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP22ama121038/Japan Agency for Medical Research and Development (AMED)
- JP22zf0127007/Japan Agency for Medical Research and Development (AMED)
- JPMJFR215T/MEXT | Japan Science and Technology Agency (JST)
- JPMJMS2023/MEXT | Japan Science and Technology Agency (JST)
- 22714181/MEXT | Japan Science and Technology Agency (JST)
LinkOut - more resources
Full Text Sources
Miscellaneous

