Mucosal adenovirus vaccine boosting elicits IgA and durably prevents XBB.1.16 infection in nonhuman primates
- PMID: 39227514
- PMCID: PMC11436372
- DOI: 10.1038/s41590-024-01951-5
Mucosal adenovirus vaccine boosting elicits IgA and durably prevents XBB.1.16 infection in nonhuman primates
Abstract
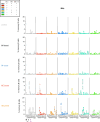
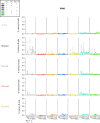
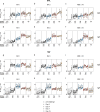
A mucosal route of vaccination could prevent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication at the site of infection and limit transmission. We compared protection against heterologous XBB.1.16 challenge in nonhuman primates (NHPs) ~5 months following intramuscular boosting with bivalent mRNA encoding WA1 and BA.5 spike proteins or mucosal boosting with a WA1-BA.5 bivalent chimpanzee adenoviral-vectored vaccine delivered by intranasal or aerosol device. NHPs boosted by either mucosal route had minimal virus replication in the nose and lungs, respectively. By contrast, protection by intramuscular mRNA was limited to the lower airways. The mucosally delivered vaccine elicited durable airway IgG and IgA responses and, unlike the intramuscular mRNA vaccine, induced spike-specific B cells in the lungs. IgG, IgA and T cell responses correlated with protection in the lungs, whereas mucosal IgA alone correlated with upper airway protection. This study highlights differential mucosal and serum correlates of protection and how mucosal vaccines can durably prevent infection against SARS-CoV-2.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
M.S.D. is a consultant for Inbios, Vir Biotechnology, Ocugen, Topspin Therapeutics, Merck, GSK and Moderna. The laboratory of M.S.D. has received unrelated funding support in sponsored research agreements from Vir Biotechnology, Emergent BioSolutions and Moderna. D.T.C. is a consultant for Ocugen, Circero, Asgard, Accession and Tome Biosciences. M.S.D., D.T.C. and I.P.D. are inventors of the ChAd-SARS-CoV-2 technology, which Washington University has licensed to Bharat Biotech and Ocugen for commercial development. M.S.S. serves on the scientific board of advisors for Moderna and Ocugen. A.R.H., P.D.K., M.R. and D.C.D. are inventors on US Patent Application 63/147,419 entitled ‘Antibodies targeting the spike protein of coronaviruses’. L.P., A.D., K.K., S.Z., A.V.R., M.G.L. and H.A. are employees of Bioqual. The other authors declare no competing interests.
Figures



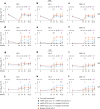
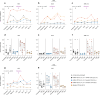



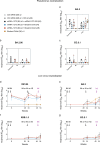

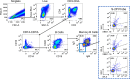


Update of
-
Mucosal Adenoviral-vectored Vaccine Boosting Durably Prevents XBB.1.16 Infection in Nonhuman Primates.bioRxiv [Preprint]. 2023 Nov 8:2023.11.06.565765. doi: 10.1101/2023.11.06.565765. bioRxiv. 2023. Update in: Nat Immunol. 2024 Oct;25(10):1913-1927. doi: 10.1038/s41590-024-01951-5. PMID: 37986823 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

