Single-cell RNA sequencing illuminates the ontogeny, conservation and diversification of cartilaginous and bony fish lymphocytes
- PMID: 39227568
- PMCID: PMC11372145
- DOI: 10.1038/s41467-024-51761-4
Single-cell RNA sequencing illuminates the ontogeny, conservation and diversification of cartilaginous and bony fish lymphocytes
Abstract
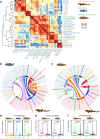
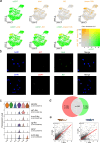
Elucidating cellular architecture and cell-type evolution across species is central to understanding immune system function and susceptibility to disease. Adaptive immunity is a shared trait of the common ancestor of cartilaginous and bony fishes. However, evolutionary features of lymphocytes in these two jawed vertebrates remain unclear. Here, we present a single-cell RNA sequencing atlas of immune cells from cartilaginous (white-spotted bamboo shark) and bony (zebrafish and Chinese tongue sole) fishes. Cross-species comparisons show that the same cell types across different species exhibit similar transcriptional profiles. In the bamboo shark, we identify a phagocytic B cell population expressing several pattern recognition receptors, as well as a T cell sub-cluster co-expressing both T and B cell markers. In contrast to a division by function in the bony fishes, we show close linkage and poor functional specialization among lymphocytes in the cartilaginous fish. Our cross-species single-cell comparison presents a resource for uncovering the origin and evolution of the gnathostome immune system.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources

