Single-cell detection of copy number changes reveals dynamic mechanisms of adaptation to antifungals in Candida albicans
- PMID: 39227665
- PMCID: PMC11524788
- DOI: 10.1038/s41564-024-01795-7
Single-cell detection of copy number changes reveals dynamic mechanisms of adaptation to antifungals in Candida albicans
Abstract
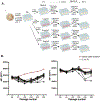
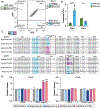
Genomic copy number changes are associated with antifungal drug resistance and virulence across diverse fungal pathogens, but the rate and dynamics of these genomic changes in the presence of antifungal drugs are unknown. Here we optimized a dual-fluorescent reporter system in the diploid pathogen Candida albicans to quantify haplotype-specific copy number variation (CNV) and loss of heterozygosity (LOH) at the single-cell level with flow cytometry. We followed the frequency and dynamics of CNV and LOH at two distinct genomic locations in the presence and absence of antifungal drugs in vitro and in a murine model of candidiasis. Copy number changes were rapid and dynamic during adaptation to fluconazole and frequently involved competing subpopulations with distinct genotypes. This study provides quantitative evidence for the rapid speed at which diverse genotypes arise and undergo dynamic population-level fluctuations during adaptation to antifungal drugs in vitro and in vivo.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures













References
-
- Pfaller MA Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med 125, S3–13 (2012). - PubMed
-
- Andes DR et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin. Infect. Dis 54, 1110–1122 (2012). - PubMed
-
- Cowen LE The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol 6, 187–198 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

