Population structure and breed identification of Chinese indigenous sheep breeds using whole genome SNPs and InDels
- PMID: 39227836
- PMCID: PMC11370120
- DOI: 10.1186/s12711-024-00927-1
Population structure and breed identification of Chinese indigenous sheep breeds using whole genome SNPs and InDels
Abstract
Background: Accurate breed identification is essential for the conservation and sustainable use of indigenous farm animal genetic resources. In this study, we evaluated the phylogenetic relationships and genomic breed compositions of 13 sheep breeds using SNP and InDel data from whole genome sequencing. The breeds included 11 Chinese indigenous and 2 foreign commercial breeds. We compared different strategies for breed identification with respect to different marker types, i.e. SNPs, InDels, and a combination of SNPs and InDels (named SIs), different breed-informative marker detection methods, and different machine learning classification methods.
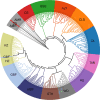
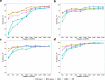
Results: Using WGS-based SNPs and InDels, we revealed the phylogenetic relationships between 11 Chinese indigenous and two foreign sheep breeds and quantified their purities through estimated genomic breed compositions. We found that the optimal strategy for identifying these breeds was the combination of DFI_union for breed-informative marker detection, which integrated the methods of Delta, Pairwise Wright's FST, and Informativeness for Assignment (namely DFI) by merging the breed-informative markers derived from the three methods, and KSR for breed assignment, which integrated the methods of K-Nearest Neighbor, Support Vector Machine, and Random Forest (namely KSR) by intersecting their results. Using SI markers improved the identification accuracy compared to using SNPs or InDels alone. We achieved accuracies over 97.5% when using at least the 1000 most breed-informative (MBI) SI markers and even 100% when using 5000 SI markers.
Conclusions: Our results provide not only an important foundation for conservation of these Chinese local sheep breeds, but also general approaches for breed identification of indigenous farm animal breeds.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Getachew T, Huson HJ, Wurzinger M, Burgstaller J, Gizaw S, Haile A, et al. Identifying highly informative genetic markers for quantification of ancestry proportions in crossbred sheep populations: Implications for choosing optimum levels of admixture. BMC Genet. 2017;18:80. 10.1186/s12863-017-0526-2 - DOI - PMC - PubMed
-
- Xu Z, Diao S, Teng J, Chen Z, Feng X, Cai X, et al. Breed identification of meat using machine learning and breed tag SNPs. Food Control. 2021;125: 107971. 10.1016/j.foodcont.2021.107971 - DOI
-
- Wang J, Lei Q, Cao D, Zhou Y, Han H, Liu W, et al. Whole genome SNPs among 8 chicken breeds enable identification of genetic signatures that underlie breed features. J Integr Agric. 2023;22:2200–12. 10.1016/j.jia.2022.11.007 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

