PanKA: Leveraging population pangenome to predict antibiotic resistance
- PMID: 39228791
- PMCID: PMC11369404
- DOI: 10.1016/j.isci.2024.110623
PanKA: Leveraging population pangenome to predict antibiotic resistance
Abstract
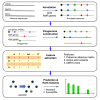
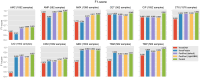
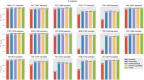
Machine learning has the potential to be a powerful tool in the fight against antimicrobial resistance (AMR), a critical global health issue. Machine learning can identify resistance mechanisms from DNA sequence data without prior knowledge. The first step in building a machine learning model is a feature extraction from sequencing data. Traditional methods like single nucleotide polymorphism (SNP) calling and k-mer counting yield numerous, often redundant features, complicating prediction and analysis. In this paper, we propose PanKA, a method using the pangenome to extract a concise set of relevant features for predicting AMR. PanKA not only enables fast model training and prediction but also improves accuracy. Applied to the Escherichia coli and Klebsiella pneumoniae bacterial species, our model is more accurate than conventional and state-of-the-art methods in predicting AMR.
Keywords: bacteriology; genomics; machine learning.
© 2024 The Author(s).
Conflict of interest statement
M.D.C., T.N., and H.A.N. are founders of AMROMICS JSC. H.S.N is a consultant to AMROMICS JSC.
Figures



References
-
- Khaledi A., Weimann A., Schniederjans M., Asgari E., Kuo T.H., Oliver A., Cabot G., Kola A., Gastmeier P., Hogardt M., et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.201910264. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

