This is a preprint.
Discovery of potent SARS-CoV-2 nsp3 macrodomain inhibitors uncovers lack of translation to cellular antiviral response
- PMID: 39229055
- PMCID: PMC11370477
- DOI: 10.1101/2024.08.19.608619
Discovery of potent SARS-CoV-2 nsp3 macrodomain inhibitors uncovers lack of translation to cellular antiviral response
Abstract
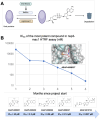
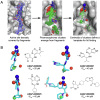
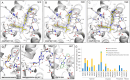
A strategy for pandemic preparedness is the development of antivirals against a wide set of viral targets with complementary mechanisms of action. SARS-CoV-2 nsp3-mac1 is a viral macrodomain with ADP-ribosylhydrolase activity, which counteracts host immune response. Targeting the virus' immunomodulatory functionality offers a differentiated strategy to inhibit SARS-CoV-2 compared to approved therapeutics, which target viral replication directly. Here we report a fragment-based lead generation campaign guided by computational approaches. We discover tool compounds which inhibit nsp3-mac1 activity at low nanomolar concentrations, and with responsive structure-activity relationships, high selectivity, and drug-like properties. Using our inhibitors, we show that inhibition of nsp3-mac1 increases ADP-ribosylation, but surprisingly does not translate to demonstrable antiviral activity in cell culture and iPSC-derived pneumocyte models. Further, no synergistic activity is observed in combination with interferon gamma, a main protease inhibitor, nor a papain-like protease inhibitor. Our results question the extent to which targeting modulation of innate immunity-driven ADP-ribosylation can influence SARS-CoV-2 replication. Moreover, these findings suggest that nsp3-mac1 might not be a suitable target for antiviral therapeutics development.
Figures




References
-
- Garnsey M. R., Robinson M. C., Nguyen L. T., Cardin R., Tillotson J., Mashalidis E., Yu A., Aschenbrenner L., Balesano A., Behzadi A., Boras B., Chang J. S., Eng H., Ephron A., Foley T., Ford K. K., Frick J. M., Gibson S., Hao L., Hurst B., Kalgutkar A. S., Korczynska M., Lengyel-Zhand Z., Gao L., Meredith H. R., Patel N. C., Polivkova J., Rai D., Rose C. R., Rothan H., Sakata S. K., Vargo T. R., Qi W., Wu H., Liu Y., Yurgelonis I., Zhang J., Zhu Y., Zhang L., Lee A. A., Discovery of SARS-CoV-2 papain-like protease (PLpro) inhibitors with efficacy in a murine infection model, bioRxiv (2024)p. 2024.01.26.577395. - PMC - PubMed
-
- Russo L. C., Tomasin R., Matos I. A., Manucci A. C., Sowa S. T., Dale K., Caldecott K. W., Lehtiö L., Schechtman D., Meotti F. C., Bruni-Cardoso A., Hoch N. C., The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 297, 101041 (2021). - PMC - PubMed
-
- Rack J. G. M., Perina D., Ahel I., Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu. Rev. Biochem. 85, 431–454 (2016). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
