MRGM: an enhanced catalog of mouse gut microbial genomes substantially broadening taxonomic and functional landscapes
- PMID: 39230075
- PMCID: PMC11376411
- DOI: 10.1080/19490976.2024.2393791
MRGM: an enhanced catalog of mouse gut microbial genomes substantially broadening taxonomic and functional landscapes
Abstract
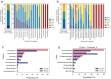
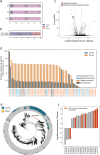
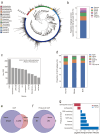
Mouse gut microbiome research is pivotal for understanding the human gut microbiome, providing insights into disease modeling, host-microbe interactions, and the dietary influence on the gut microbiome. To enhance the translational value of mouse gut microbiome studies, we need detailed and high-quality catalogs of mouse gut microbial genomes. We introduce the Mouse Reference Gut Microbiome (MRGM), a comprehensive catalog with 42,245 non-redundant mouse gut bacterial genomes across 1,524 species. MRGM marks a 40% increase in the known taxonomic diversity of mouse gut microbes, capturing previously underrepresented lineages through refined genome quality assessment techniques. MRGM not only broadens the taxonomic landscape but also enriches the functional landscape of the mouse gut microbiome. Using deep learning, we have elevated the Gene Ontology annotation rate for mouse gut microbial proteins from 3.2% with orthology to 60%, marking an over 18-fold increase. MRGM supports both DNA- and marker-based taxonomic profiling by providing custom databases, surpassing previous catalogs in performance. Finally, taxonomic and functional comparisons between human and mouse gut microbiota reveal diet-driven divergences in their taxonomic composition and functional enrichment. Overall, our study highlights the value of high-quality microbial genome catalogs in advancing our understanding of the co-evolution between gut microbes and their host.
Keywords: Gut microbiome; metagenome-assembled genomes; mouse gut microbiome; reference microbiome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




Update of
-
MRGM: An enhanced catalog of mouse gut microbial genomes substantially broadening taxonomic and functional landscapes.bioRxiv [Preprint]. 2024 Aug 12:2024.08.12.607507. doi: 10.1101/2024.08.12.607507. bioRxiv. 2024. Update in: Gut Microbes. 2024 Jan-Dec;16(1):2393791. doi: 10.1080/19490976.2024.2393791. PMID: 39211244 Free PMC article. Updated. Preprint.
Similar articles
-
MRGM: An enhanced catalog of mouse gut microbial genomes substantially broadening taxonomic and functional landscapes.bioRxiv [Preprint]. 2024 Aug 12:2024.08.12.607507. doi: 10.1101/2024.08.12.607507. bioRxiv. 2024. Update in: Gut Microbes. 2024 Jan-Dec;16(1):2393791. doi: 10.1080/19490976.2024.2393791. PMID: 39211244 Free PMC article. Updated. Preprint.
-
An Expanded Gene Catalog of Mouse Gut Metagenomes.mSphere. 2021 Feb 24;6(1):e01119-20. doi: 10.1128/mSphere.01119-20. mSphere. 2021. PMID: 33627510 Free PMC article.
-
Human reference gut microbiome catalog including newly assembled genomes from under-represented Asian metagenomes.Genome Med. 2021 Aug 27;13(1):134. doi: 10.1186/s13073-021-00950-7. Genome Med. 2021. PMID: 34446072 Free PMC article.
-
A review on recent taxonomic updates of gut bacteria associated with social bees, with a curated genomic reference database.Insect Sci. 2025 Feb;32(1):2-23. doi: 10.1111/1744-7917.13365. Epub 2024 Apr 9. Insect Sci. 2025. PMID: 38594229 Review.
-
Genome-resolved metagenomics: a game changer for microbiome medicine.Exp Mol Med. 2024 Jul;56(7):1501-1512. doi: 10.1038/s12276-024-01262-7. Epub 2024 Jul 1. Exp Mol Med. 2024. PMID: 38945961 Free PMC article. Review.
Cited by
-
A conserved pilin from uncultured gut bacterial clade TANB77 enhances cancer immunotherapy.Nat Commun. 2024 Dec 27;15(1):10726. doi: 10.1038/s41467-024-55388-3. Nat Commun. 2024. PMID: 39730328 Free PMC article.
References
-
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi:10.1038/s41591-019-0406-6. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
