Three families of CD4-induced antibodies are associated with the capacity of plasma from people living with HIV to mediate ADCC in the presence of CD4-mimetics
- PMID: 39230306
- PMCID: PMC11495032
- DOI: 10.1128/jvi.00960-24
Three families of CD4-induced antibodies are associated with the capacity of plasma from people living with HIV to mediate ADCC in the presence of CD4-mimetics
Abstract
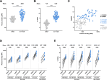
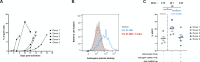
CD4-mimetics (CD4mcs) are small molecule compounds that mimic the interaction of the CD4 receptor with HIV-1 envelope glycoproteins (Env). Env from primary viruses normally samples a "closed" conformation that occludes epitopes recognized by CD4-induced (CD4i) non-neutralizing antibodies (nnAbs). CD4mcs induce conformational changes on Env resulting in the exposure of these otherwise inaccessible epitopes. Here, we evaluated the capacity of plasma from a cohort of 50 people living with HIV to recognize HIV-1-infected cells and eliminate them by antibody-dependent cellular cytotoxicity (ADCC) in the presence of a potent indoline CD4mc. We observed a marked heterogeneity among plasma samples. By measuring the levels of different families of CD4i Abs, we found that the levels of anti-cluster A, anti-coreceptor binding site, and anti-gp41 cluster I antibodies are responsible for plasma-mediated ADCC in the presence of CD4mc.
Importance: There are several reasons that make it difficult to target the HIV reservoir. One of them is the capacity of infected cells to prevent the recognition of HIV-1 envelope glycoproteins (Env) by commonly elicited antibodies in people living with HIV. Small CD4-mimetic compounds expose otherwise occluded Env epitopes, thus enabling their recognition by non-neutralizing antibodies (nnAbs). A better understanding of the contribution of these antibodies to eliminate infected cells in the presence of CD4mc could lead to the development of therapeutic cure strategies.
Keywords: ADCC; CD4 mimetics; Env; HIV cure; HIV-1; cluster A region; co-receptor binding site; gp41 cluster I; non-neutralizing antibodies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Update of
-
Three families of CD4-induced antibodies are associated with the capacity of plasma from people living with HIV to mediate ADCC in presence of CD4-mimetics.medRxiv [Preprint]. 2024 Jun 4:2024.06.02.24308281. doi: 10.1101/2024.06.02.24308281. medRxiv. 2024. Update in: J Virol. 2024 Oct 22;98(10):e0096024. doi: 10.1128/jvi.00960-24. PMID: 38883797 Free PMC article. Updated. Preprint.
Similar articles
-
The combination of three CD4-induced antibodies targeting highly conserved Env regions with a small CD4-mimetic achieves potent ADCC activity.J Virol. 2024 Oct 22;98(10):e0101624. doi: 10.1128/jvi.01016-24. Epub 2024 Sep 9. J Virol. 2024. PMID: 39248460 Free PMC article.
-
Three families of CD4-induced antibodies are associated with the capacity of plasma from people living with HIV to mediate ADCC in presence of CD4-mimetics.medRxiv [Preprint]. 2024 Jun 4:2024.06.02.24308281. doi: 10.1101/2024.06.02.24308281. medRxiv. 2024. Update in: J Virol. 2024 Oct 22;98(10):e0096024. doi: 10.1128/jvi.00960-24. PMID: 38883797 Free PMC article. Updated. Preprint.
-
Co-receptor Binding Site Antibodies Enable CD4-Mimetics to Expose Conserved Anti-cluster A ADCC Epitopes on HIV-1 Envelope Glycoproteins.EBioMedicine. 2016 Oct;12:208-218. doi: 10.1016/j.ebiom.2016.09.004. Epub 2016 Sep 9. EBioMedicine. 2016. PMID: 27633463 Free PMC article.
-
Unlocking HIV-1 Env: implications for antibody attack.AIDS Res Ther. 2017 Sep 12;14(1):42. doi: 10.1186/s12981-017-0168-5. AIDS Res Ther. 2017. PMID: 28893275 Free PMC article. Review.
-
Conformation-Dependent Interactions Between HIV-1 Envelope Glycoproteins and Broadly Neutralizing Antibodies.AIDS Res Hum Retroviruses. 2018 Sep;34(9):794-803. doi: 10.1089/AID.2018.0102. Epub 2018 Jul 17. AIDS Res Hum Retroviruses. 2018. PMID: 29905080 Review.
Cited by
-
CD4 T cell counts are inversely correlated with anti-gp120 cluster A antibodies in antiretroviral therapy-treated PLWH.EBioMedicine. 2025 Aug;118:105856. doi: 10.1016/j.ebiom.2025.105856. Epub 2025 Jul 19. EBioMedicine. 2025. PMID: 40684476 Free PMC article.
-
Safety, pharmacokinetics, and biological activity of CD4-mimetic BNM-III-170 in SHIV-infected rhesus macaques.J Virol. 2025 May 20;99(5):e0006225. doi: 10.1128/jvi.00062-25. Epub 2025 Apr 7. J Virol. 2025. PMID: 40192306 Free PMC article.
-
The combination of three CD4-induced antibodies targeting highly conserved Env regions with a small CD4-mimetic achieves potent ADCC activity.bioRxiv [Preprint]. 2024 Jun 7:2024.06.07.597978. doi: 10.1101/2024.06.07.597978. bioRxiv. 2024. Update in: J Virol. 2024 Oct 22;98(10):e0101624. doi: 10.1128/jvi.01016-24. PMID: 38895270 Free PMC article. Updated. Preprint.
-
Optimization of a Piperidine CD4-Mimetic Scaffold Sensitizing HIV-1 Infected Cells to Antibody-Dependent Cellular Cytotoxicity.ACS Med Chem Lett. 2024 Oct 28;15(11):1961-1969. doi: 10.1021/acsmedchemlett.4c00403. eCollection 2024 Nov 14. ACS Med Chem Lett. 2024. PMID: 39563795
-
The asymmetric opening of HIV-1 Env by a potent CD4 mimetic enables anti-coreceptor binding site antibodies to mediate ADCC.bioRxiv [Preprint]. 2024 Aug 27:2024.08.27.609961. doi: 10.1101/2024.08.27.609961. bioRxiv. 2024. PMID: 39253431 Free PMC article. Preprint.
References
-
- Ma X, Lu M, Gorman J, Terry DS, Hong X, Zhou Z, Zhao H, Altman RB, Arthos J, Blanchard SC, Kwong PD, Munro JB, Mothes W. 2018. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. Elife 7:e34271. doi:10.7554/eLife.34271 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- master fellowship/Canadian Government | Canadian Institutes of Health Research (CIHR)
- R01 AI148379/AI/NIAID NIH HHS/United States
- R01 AI176531/AI/NIAID NIH HHS/United States
- R01 AI129769/AI/NIAID NIH HHS/United States
- R01 AI150322/AI/NIAID NIH HHS/United States
- AI148379,AI150322,AI176531,AI164562/HHS | National Institutes of Health (NIH)
- 422148,451304/Canadian Government | Canadian Institutes of Health Research (CIHR)
- Elevation/Mitacs (Mitacs Canada)
- doctoral fellowship/Canadian Government | Canadian Institutes of Health Research (CIHR)
- R01 AI186809/AI/NIAID NIH HHS/United States
- R01 AI174908/AI/NIAID NIH HHS/United States
- AI129769,AI174908, AI186809/HHS | National Institutes of Health (NIH)
- 41027/Canada Foundation for Innovation (CFI)
- UM1 AI164562/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

