Structure-based design of a soluble human cytomegalovirus glycoprotein B antigen stabilized in a prefusion-like conformation
- PMID: 39231203
- PMCID: PMC11406251
- DOI: 10.1073/pnas.2404250121
Structure-based design of a soluble human cytomegalovirus glycoprotein B antigen stabilized in a prefusion-like conformation
Abstract
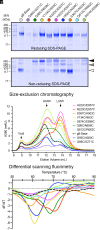
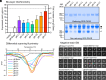
Human cytomegalovirus (HCMV) glycoprotein B (gB) is a class III membrane fusion protein required for viral entry. HCMV vaccine candidates containing gB have demonstrated moderate clinical efficacy, but no HCMV vaccine has been approved. Here, we used structure-based design to identify and characterize amino acid substitutions that stabilize gB in its metastable prefusion conformation. One variant containing two engineered interprotomer disulfide bonds and two cavity-filling substitutions (gB-C7), displayed increased expression and thermostability. A 2.8 Å resolution cryoelectron microscopy structure shows that gB-C7 adopts a prefusion-like conformation, revealing additional structural elements at the membrane-distal apex. Unlike previous observations for several class I viral fusion proteins, mice immunized with postfusion or prefusion-stabilized forms of soluble gB protein displayed similar neutralizing antibody titers, here specifically against an HCMV laboratory strain on fibroblasts. Collectively, these results identify initial strategies to stabilize class III viral fusion proteins and provide tools to probe gB-directed antibody responses.
Keywords: cryo-EM; cytomegalovirus; herpesvirus; vaccine.
Conflict of interest statement
Competing interests statement:M.R.S., P.O.B., A.R.R., J.A.G., R.S.M., L.Z., N.V.J., C.-L.H., and J.S.M. are inventors on a patent application entitled “Prefusion-Stabilized CMV gB Proteins” (PCT/US2023/073369). J.D.C., D.Y., and M.J.B. are employees of Dynavax and hold Dynavax stock. The other authors declare that they have no competing interests.
Figures



References
-
- Anonymous, “Babies born with congenital CMV” in Cytomegalovirus (CMV) and Congenital CMV Infection (Centers for Disease Control and Prevention, 2022).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

