BioID-based intact cell interactome of the Kv1.3 potassium channel identifies a Kv1.3-STAT3-p53 cellular signaling pathway
- PMID: 39231216
- PMCID: PMC11373599
- DOI: 10.1126/sciadv.adn9361
BioID-based intact cell interactome of the Kv1.3 potassium channel identifies a Kv1.3-STAT3-p53 cellular signaling pathway
Abstract
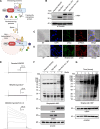
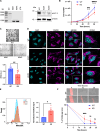
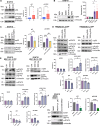
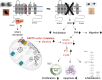
Kv1.3 is a multifunctional potassium channel implicated in multiple pathologies, including cancer. However, how it is involved in disease progression is not fully clear. We interrogated the interactome of Kv1.3 in intact cells using BioID proximity labeling, revealing that Kv1.3 interacts with STAT3- and p53-linked pathways. To prove the relevance of Kv1.3 and of its interactome in the context of tumorigenesis, we generated stable melanoma clones, in which ablation of Kv1.3 remodeled gene expression, reduced proliferation and colony formation, yielded fourfold smaller tumors, and decreased metastasis in vivo in comparison to WT cells. Kv1.3 deletion or pharmacological inhibition of mitochondrial Kv1.3 increased mitochondrial Reactive Oxygen Species release, decreased STAT3 phosphorylation, stabilized the p53 tumor suppressor, promoted metabolic switch, and altered the expression of several BioID-identified Kv1.3-networking proteins in tumor tissues. Collectively, our work revealed the tumor-promoting Kv1.3-interactome landscape, thus opening the way to target Kv1.3 not only as an ion-conducting entity but also as a signaling hub.
Figures




References
-
- Pardo L. A., Stuhmer W., The roles of K+ channels in cancer. Nat. Rev. Cancer 14, 39–48 (2014). - PubMed
-
- Perez-Verdaguer M., Capera J., Serrano-Novillo C., Estadella I., Sastre D., Felipe A., The voltage-gated potassium channel Kv1.3 is a promising multitherapeutic target against human pathologies. Expert Opin. Ther. Targets 20, 577–591 (2016). - PubMed
-
- Sarkar S., Nguyen H. M., Malovic E., Luo J., Langley M., Palanisamy B. N., Singh N., Manne S., Neal M., Gabrielle M., Abdalla A., Anantharam P., Rokad D., Panicker N., Singh V., Ay M., Charli A., Harischandra D., Jin L. W., Jin H., Rangaraju S., Anantharam V., Wulff H., Kanthasamy A. G., Kv1.3 modulates neuroinflammation and neurodegeneration in Parkinson’s disease. J. Clin. Invest. 130, 4195–4212 (2020). - PMC - PubMed
-
- Beeton C., Wulff H., Standifer N. E., Azam P., Mullen K. M., Pennington M. W., Kolski-Andreaco A., Wei E., Grino A., Counts D. R., Wang P. H., LeeHealey C. J., Andrews B. S., Sankaranarayanan A., Homerick D., Roeck W. W., Tehranzadeh J., Stanhope K. L., Zimin P., Havel P. J., Griffey S., Knaus H. G., Nepom G. T., Gutman G. A., Calabresi P. A., Chandy K. G., Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc. Natl. Acad. Sci. U.S.A 103, 17414–17419 (2006). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

