Effector memory-type regulatory T cells display phenotypic and functional instability
- PMID: 39231218
- PMCID: PMC11421655
- DOI: 10.1126/sciadv.adn3470
Effector memory-type regulatory T cells display phenotypic and functional instability
Abstract
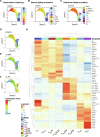
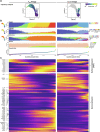
Regulatory T cells (Treg cells) hold promise for sustainable therapy of immune disorders. Recent advancements in chimeric antigen receptor development and genome editing aim to enhance the specificity and function of Treg cells. However, impurities and functional instability pose challenges for the development of safe gene-edited Treg cell products. Here, we examined different Treg cell subsets regarding their fate, epigenomic stability, transcriptomes, T cell receptor repertoires, and function ex vivo and after manufacturing. Each Treg cell subset displayed distinct features, including lineage stability, epigenomics, surface markers, T cell receptor diversity, and transcriptomics. Earlier-differentiated memory Treg cell populations, including a hitherto unidentified naïve-like memory Treg cell subset, outperformed late-differentiated effector memory-like Treg cells in regulatory function, proliferative capacity, and epigenomic stability. High yields of stable, functional Treg cell products could be achieved by depleting the small effector memory-like Treg cell subset before manufacturing. Considering Treg cell subset composition appears critical to maintain lineage stability in the final cell product.
Figures






References
-
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M., Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). - PubMed
-
- Roemhild A., Otto N. M., Moll G., Abou-El-Enein M., Kaiser D., Bold G., Schachtner T., Choi M., Oellinger R., Landwehr-Kenzel S., Juerchott K., Sawitzki B., Giesler C., Sefrin A., Beier C., Wagner D. L., Schlickeiser S., Streitz M., Schmueck-Henneresse M., Amini L., Stervbo U., Babel N., Volk H.-D., Reinke P., Regulatory T cells for minimising immune suppression in kidney transplantation: Phase I/IIa clinical trial. BMJ 371, m3734 (2020). - PMC - PubMed
-
- Landwehr-Kenzel S., Mueller-Jensen L., Kuehl J. S., Abou-El-Enein M., Hoffmann H., Muench S., Kaiser D., Roemhild A., von Bernuth H., Voeller M., Schmueck-Henneresse M., Gruhn B., Stervbo U., Babel N., Volk H. D., Reinke P., Adoptive transfer of ex vivo expanded regulatory T-cells improves immune cell engraftment and therapy-refractory chronic GvHD. Mol. Ther. 30, 2298–2314 (2022). - PMC - PubMed
-
- Oo Y. H., Ackrill S., Cole R., Jenkins L., Anderson P., Jeffery H. C., Jones N., Jeffery L. E., Lutz P., Wawman R. E., Athwal A. K., Thompson J., Gray J., Guo K., Barton D., Hirschfield G. M., Wong T., Guest P., Adams D. H., Liver homing of clinical grade Tregs after therapeutic infusion in patients with autoimmune hepatitis. JHEP Rep. 1, 286–296 (2019). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

