Broad Adaptability of Coronavirus Adhesion Revealed from the Complementary Surface Affinity of Membrane and Spikes
- PMID: 39231361
- PMCID: PMC11538687
- DOI: 10.1002/advs.202404186
Broad Adaptability of Coronavirus Adhesion Revealed from the Complementary Surface Affinity of Membrane and Spikes
Abstract
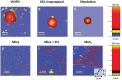
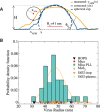
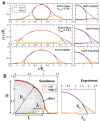
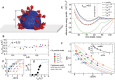
Coronavirus stands for a large family of viruses characterized by protruding spikes surrounding a lipidic membrane adorned with proteins. The present study explores the adhesion of transmissible gastroenteritis coronavirus (TGEV) particles on a variety of reference solid surfaces that emulate typical virus-surface interactions. Atomic force microscopy informs about trapping effectivity and the shape of the virus envelope on each surface, revealing that the deformation of TGEV particles spans from 20% to 50% in diameter. Given this large deformation range, experimental Langmuir isotherms convey an unexpectedly moderate variation in the adsorption-free energy, indicating a viral adhesion adaptability which goes beyond the membrane. The combination of an extended Helfrich theory and coarse-grained simulations reveals that, in fact, the envelope and the spikes present complementary adsorption affinities. While strong membrane-surface interaction lead to highly deformed TGEV particles, surfaces with strong spike attraction yield smaller deformations with similar or even larger adsorption-free energies.
Keywords: atomic‐force‐microscopy; coarse‐graining models; coronavirus; elastic theory; surface‐affinity.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus.Virol J. 2011 Sep 12;8:435. doi: 10.1186/1743-422X-8-435. Virol J. 2011. PMID: 21910859 Free PMC article.
-
The coronavirus transmissible gastroenteritis virus causes infection after receptor-mediated endocytosis and acid-dependent fusion with an intracellular compartment.J Virol. 1998 Jan;72(1):527-34. doi: 10.1128/JVI.72.1.527-534.1998. J Virol. 1998. PMID: 9420255 Free PMC article.
-
Antiviral and virucidal effects of curcumin on transmissible gastroenteritis virus in vitro.J Gen Virol. 2020 Oct;101(10):1079-1084. doi: 10.1099/jgv.0.001466. Epub 2020 Jul 16. J Gen Virol. 2020. PMID: 32677610
-
An overview of immunological and genetic methods for detecting swine coronaviruses, transmissible gastroenteritis virus, and porcine respiratory coronavirus in tissues.Adv Exp Med Biol. 1997;412:37-46. doi: 10.1007/978-1-4899-1828-4_4. Adv Exp Med Biol. 1997. PMID: 9191988 Review.
-
Immunity to transmissible gastroenteritis virus and porcine respiratory coronavirus infections in swine.Vet Immunol Immunopathol. 1994 Oct;43(1-3):89-97. doi: 10.1016/0165-2427(94)90124-4. Vet Immunol Immunopathol. 1994. PMID: 7856068 Free PMC article. Review.
Cited by
-
2Danalysis: A toolbox for analysis of lipid membranes and biopolymers in two-dimensional space.bioRxiv [Preprint]. 2025 Mar 2:2025.02.27.640563. doi: 10.1101/2025.02.27.640563. bioRxiv. 2025. Update in: Biophys J. 2025 May 30:S0006-3495(25)00321-2. doi: 10.1016/j.bpj.2025.05.026. PMID: 40060461 Free PMC article. Updated. Preprint.
References
-
- Zhang G., Cong Y., Liu F.‐L., Sun J., Zhang J., Cao G., Zhou L., Yang W., Song Q., Wang F., Liu K., Qu J., Wang J., He M., Feng S., Baimanov D., Xu W., Luo R.‐H., Long X.‐Y., Liao S., Fan Y., Li Y.‐F., Li B., Shao X., Wang G., Fang L., Wang H., Yu X.‐F., Chang Y.‐Z., Zhao Y., et al., Nat. Nanotechnol. 2022, 17, 993. - PubMed
-
- Zeng C., Hernando‐Pérez M., Dragnea B., Ma X., van der Schoot P., Zandi R., Phys. Rev. Lett. 2017, 119, 038102. - PubMed
-
- Lipowsky R., Seifert U., Mol. Cryst. Liq. Cryst. 1991, 202, 17.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
